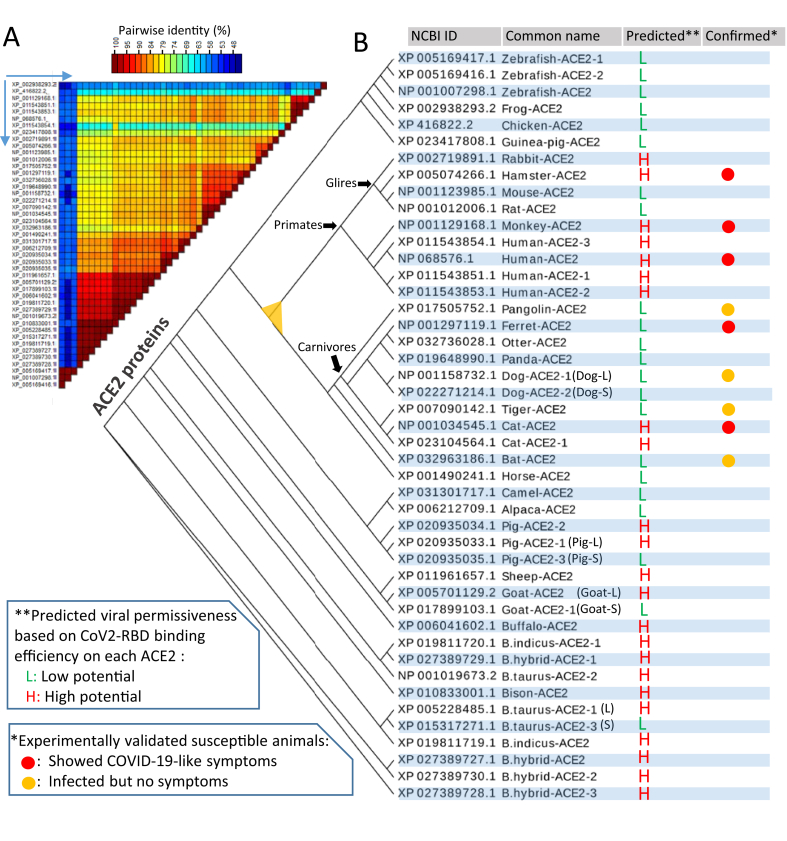
Figure 1.
Incongruence of predicted SARS-CoV2 susceptibility to experimental data in selected major livestock and reference animal species. Protein sequences of angiotensin converting enzyme 2 (ACE2) orthologs in different species, and transcript variants identified within some species, were extracted from NCBI (and Ensembl) Gene databases and verified using relevant RNA-Seq expression. (A) Sequence alignment was performed with ClustalW and pairwise identity (%) was calculated and visualized using SDT1.2. The reciprocal proteins for comparison on the horizontal are the same order as the vertical. Only NCBI Accession numbers are listed, see the common names in (B). (B) The phylogenic tree of major identified ACE2 orthologs/variants from different species was built with a Neighbor-joining approach and visualized using an EvoView program under default parameter setting. The prediction of SARS-CoV2 susceptibility is based on the sequence similarity of each ACE2 to human ACE2 in the S-RBD binding region and simulated using a published human ACE2-RBD structure (6M0J) and refers to two recent publications using similar procedures but different structural models [27, 28]. Compared with the currently available experimental data, incongruence of the predicted SARS-CoV2 susceptibility is clearly demonstrated in pangolin, ferret, tiger, cat and horseshoe bat, indicating that some other factors besides ACE2-RBD affinity should be considered.