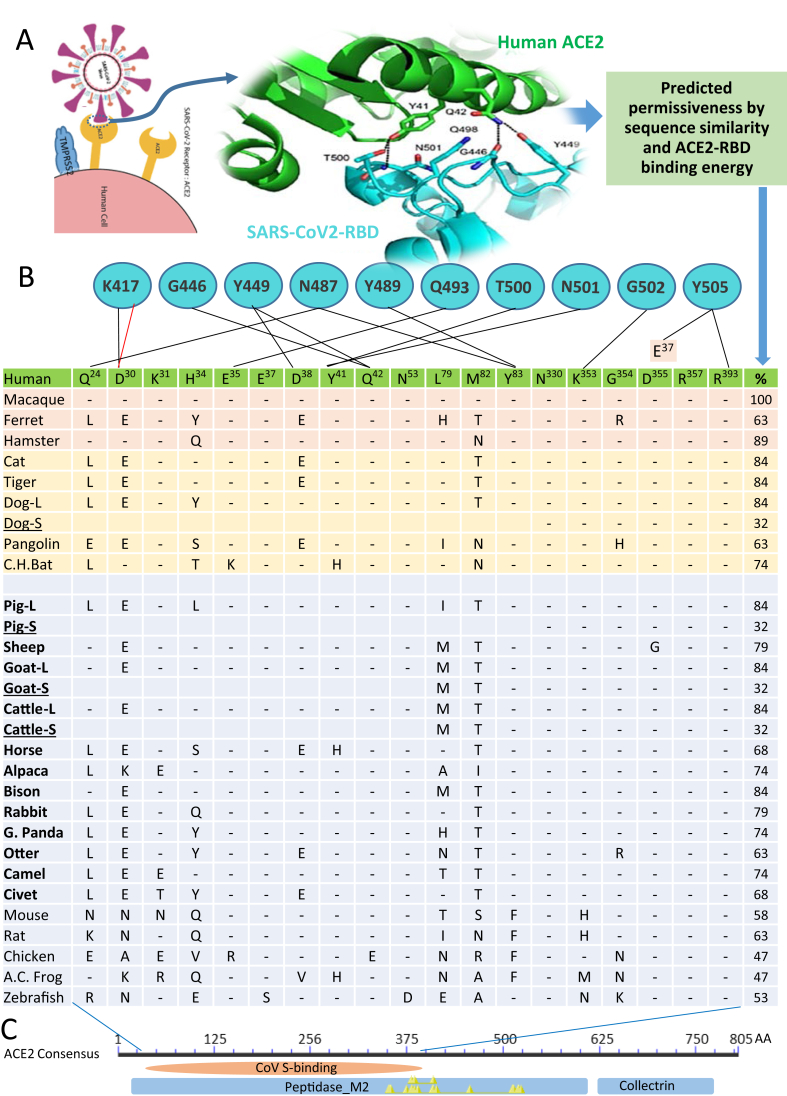
Figure 2.
Prediction of SARS-CoV2 susceptibility in major livestock species based on the conservation of key interacting residues and binding capacity between the viral spike (S) protein on the host ACE2 receptor. (A) SARS-CoV-2 uses the cell receptor, angiotensin-converting enzyme 2 (ACE2) for entry and the serine protease TMPRSS2 and furin for S protein priming. (B) As TMPRSS2 is broadly expressed and active with a furin-like cleavage activity, the affinity adaption of the S receptor binding domain (RBD) and ACE2 receptor determines the viral permissiveness. The contacting residues of human ACE2 (a distance cutoff 4.5 Å) at the SARS-CoV-2 RBD/ACE2 interfaces are shown, and the contacting network involves at least 19 residues in ACE2 (listed in the Table cells and referred to the aligned residual positions in human ACE2) and 10 residues in the SARS-CoV-2 RBD (blue circles with residue labels), which are listed and connected with black lines (indicating hydrogen bonds) and red line (represents salt-bridge interaction). The cross-species residual identity (%) of these interacting residues in ACE2 are listed in a broad range (32–100%) [26, 27, 28]. (C) We also detected several short ACE2 isoforms (underlined) in the domestic animals including dog, pig, goat and cattle, which have an N-terminal truncation spanning 10–13 key residues in the contacting network to S-RBD but keeping the enzyme active sites (indicated by Yellow triangles), thus resulting in little engagement by the viral S protein and predicting an unexpected evolutionary advantage for relieving potential COVID-19 risk caused by the viral engagement and functional distortion on the classical long ACE2 isoforms in these animal species. The NCBI Accession Numbers of the ACE2 orthologs are listed as in Figure 1.