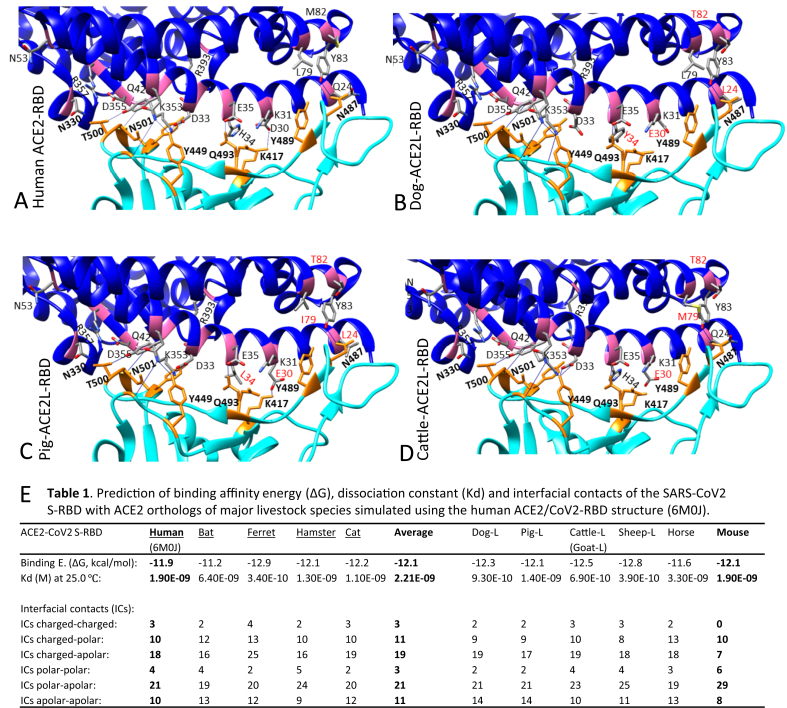
Figure 3.
Binding capacity of SARS-CoV2 spike protein (S) and its cell receptors (ACE2) from different animal species. (A) Structure of the receptor-binding domain (RBD) of S from SARS-CoV2 (cyan) bound to human angiotensin-converting enzyme 2 (ACE2) (blue). Most residues involved in binding are highlighted as magenta (ACE2) or orange (S) sticks and labeled as one-letter amino-acid codes plus residual numbers in bold or regular font respectively for S or ACE2 residues. The dotted/blue lines indicate intermolecular salt bridge or hydrogen bonds between interacting residues (generated and visualized with UCSF Chimera and Pymol from Protein Data Bank File 6M0J). (B) to (D) RBD interaction with the simulated structures of ACE2 long isoforms from the dog, pig and cattle, respectively. Amino acid exchanges in ACE2 from another species compared with human ACE2 are highlighted in red. (E) Prediction of binding affinity energy (ΔG), dissociation constant (Kd) and interfacial contacts of the SARS-CoV2 S-RBD with ACE2 orthologs of major livestock species. Most domestic animals ACE2 including that from mouse and rat (species known not to be susceptible to SARS-CoV2) have a binding affinity (ΔG) at -11.2 to -12.8 kcal/mol that is within the range (11.2–12.9 kcal/mol) between the RBD and the ACE2 from the known susceptible species (underlined in the left part of the table), indicating that some other factors, especially those from genetic divergence and natural immunity, contribute to the SARS-CoV2 susceptibility of different animal species.