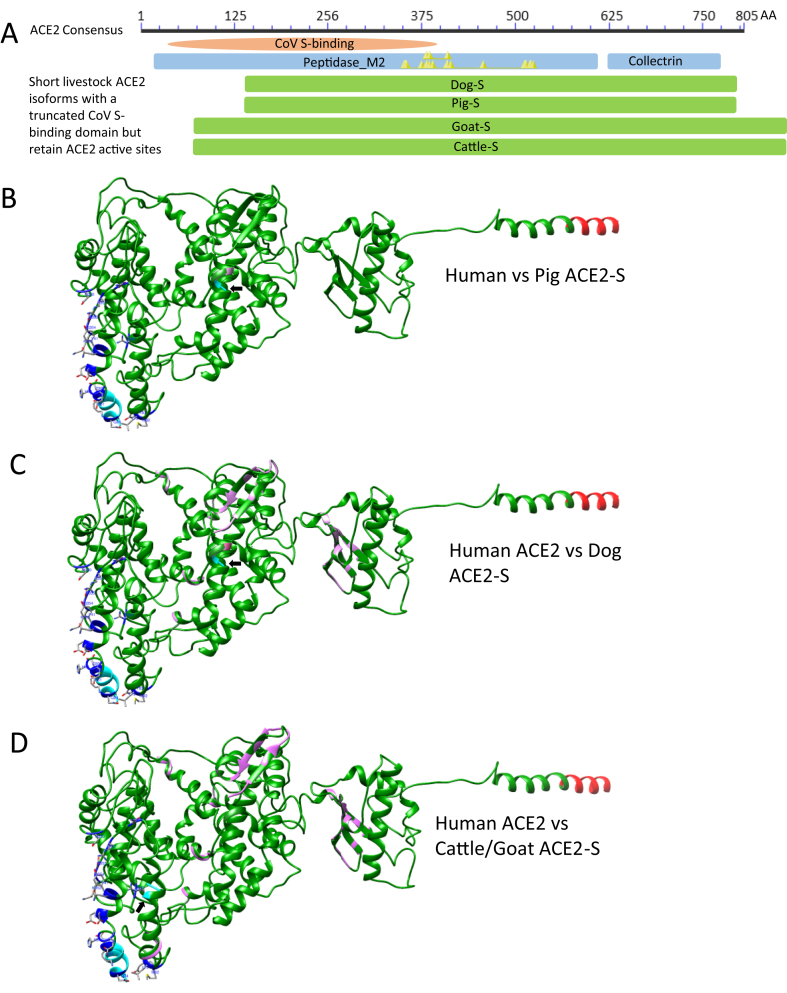
Figure 4.
Detection of several short ACE2 isoforms (ACE2-S) in the domestic animals including dog, pig, goat and cattle. (A) In contrast to most splicing isoforms such as in cats and humans, which share a common proximal promoter and encode ACE2 proteins with similar sequences containing all 19 key RBD-interacting residues, these short ACE2-S isoforms in domestic animals truncate for 71 (Cattle/Goat ACE2-S) or 132 (Dog/Pig ACE2-S) residues at their N-termini compared with human ACE2 or the long ACE2 isoforms in these species, thus destroying 10–13 key residues in the contacting network to S-RBD but retaining all enzyme active sites (Yellow triangles in the blue ACE2 domain bar). This results in little chance to be engaged by the viral S protein binding and predicts an unexpected evolutionary advantage to relieve potential COVID-19 risk caused by the viral engagement and functional distortion on the classical long ACE2 isoforms in these animal species. (B), (C) and (D) Paired structural comparison between the human ACE2 structure (6M17) with each simulated ACE2-S structure from pig (B), dog (C) and cattle/goat (D). Human ACE2 structure are in green, and each compared animal ACE2-S structure in magenta. The N-terminal residues of both compared structures are in cyan (arrows indicating N-termini of the ACE2-S isoforms) and shared C-termini are in red. The 19 key S-interacting residues in human ACE2 are shown in blue sticks. In general, all ACE2-S orthologs, particular the porcine, show high structural similarity to the human ACE2 except the N-terminal truncations.