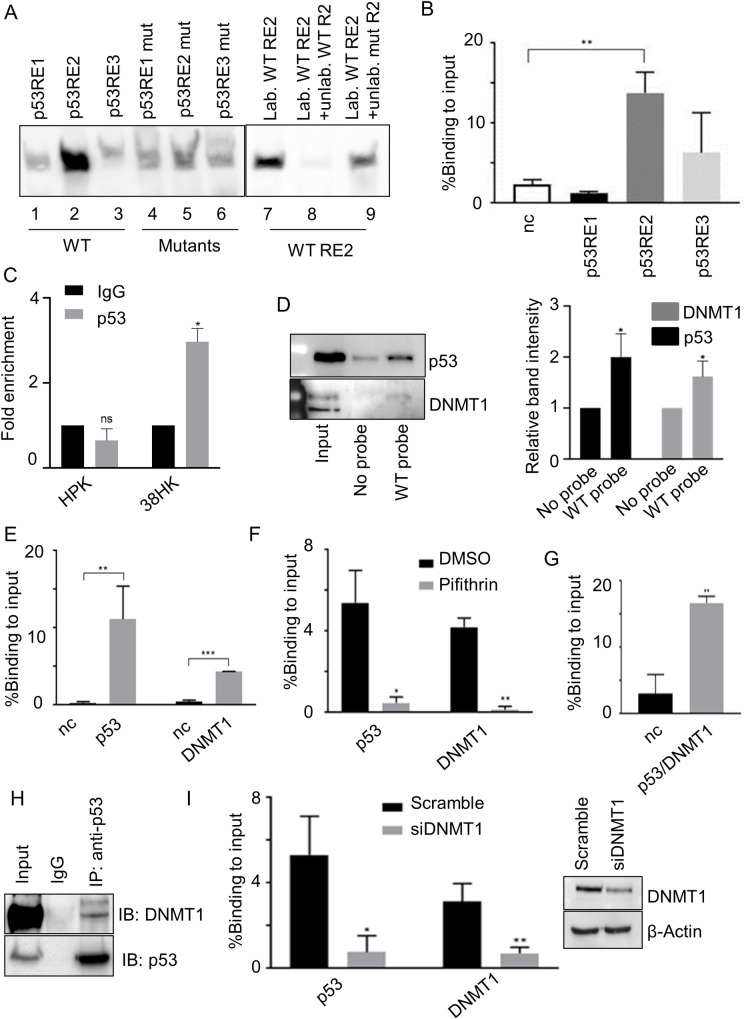
Fig 2. p53 and DNMT1 form a complex that is recruited to the ITGA1 promoter.
(A) Electromobility shift assay performed with 38HK nuclear protein extracts and biotinylated probes containing p53RE WT or mutated sequences. Probes were incubated and cross-linked with protein extracts. Unlabeled WT or mutant p53RE2 probes were used as a control. Images shown are representative examples of 2 different experiments. (B) 38HK were cross-linked and chromatin was processed for ChIP using p53 antibody. Results were analyzed by qPCR with primers spanning p53RE1, p53RE2, p53RE3, or the intergenic region of chromosome 22 as a negative control (nc). Error bars represent standard deviations of 3 independent experiments performed in triplicate. **, p<0.01. (C) HKs or 38HK were cross-linked and chromatin was processed for ChIP using p53 or IgG antibodies. Results were analyzed by qPCR using primers spanning for p53 REs of the ITGA1 promoter and normalized to IgG enrichment (negative control). Error bars represent the standard deviation of 2 independent experiments performed in 2 different HKs donors. *, p<0.05, ns, not significant. (D) Cell lysate was incubated with WT biotinylated probe containing p53 REs of the ITGA1 promoter. Incubation without a probe was used as a control. DNA-associated proteins were recovered by precipitation with streptavidin beads and analyzed by IB. Images shown are representative examples of 3 independent experiments. Signals of 3 different IBs were quantified with Image Lab software (right panel). Data shown are the means of 3 independent experiments. *, p<0.05. (E) Chromatin from 38HK was processed for ChIP experiments using p53 or DNMT1 antibodies. Results were obtained by qPCR with primers spanning p53RE2 or the intergenic region of chromosome 22 (nc). Error bars indicate standard deviations from 3 independent experiments performed in duplicate. **, p<0.01; ***, p<0.001. (F) 38HK were cultured in medium containing cyclic pifithrin-α hydrobromide or DMSO as a control. Chromatin was processed for ChIP using p53 or DNMT1 antibodies. Results were obtained by qPCR using primers spanning p53RE2. Data shown are the means of 2 independent experiments performed in triplicate. *, p<0.05, **, p<0.01. (G) Chromatin was processed for a ChIP-reChIP assay in which p53-immunoprecipitated DNA was re-immunoprecipitated by DNMT1. Enrichment of p53RE2 or the intergenic region of chromosome 22 (nc) was obtained by qPCR. Data shown are the means of 3 independent experiments performed in triplicate. **, p<0.01. (H) Nuclear protein extracts from 38HK were processed for IP. Agarose beads were conjugated with IgG or p53 antibodies. Conjugated beads were incubated with protein lysate overnight. IgG was used as a control. Results were obtained by IB using the indicated antibodies. (I) 38HK were transfected with DNMT1 siRNA or control siRNA (Scramble). After 72 h, a ChIP assay was performed with p53 or DNMT1 antibodies. Results were obtained by qPCR using p53RE2 primers. Error bars represent standard deviations from 3 independent experiments. *, p<0.05; **, p<0.01. DNMT1 protein levels in different cells were determined by IB with the indicated antibodies (right panel).