Highlights
-
•
Trypanosoma vivax strains exhibit different virulence and pathogenicity patterns.
-
•
TvMT1 strain showed low virulence and high pathogenicity.
-
•
TvLIEM176 strain showed high virulence and moderate pathogenicity.
-
•
Protein expression varies in high virulence/moderate pathogenicity strain vs low virulence/high pathogenicity strain.
Keywords: Trypanosoma vivax, Virulence, Pathogenicity, Proteomics, Biomarkers
Abstract
Cattle trypanosomosis caused by Trypanosoma vivax is a widely distributed disease in Africa and Latin America. It causes significant losses in the livestock industry and is characterized by fluctuating parasitemia, anemia, fever, lethargy, and weight loss. In this study we evaluated the virulence (capacity to multiply inside the host and to modulate the host response) and pathogenicity (ability to produce disease and/or mortality) patterns of two T. vivax strains (TvMT1 and TvLIEM176) in experimentally-infected sheep and determined the proteins differentially expressed in the proteomes of these two strains. Hematological and clinical parameters were monitored in experimentally-infected versus non-infected sheep for 60 days. All the infected animals developed discernable parasitemia at 3 days post-infection (dpi), and the first parasitemia peak was observed at 6 dpi. The maximum average value of parasitemia was 1.3 × 107 (95% CI, 7.9 × 105–2 × 108) parasites/ml in TvLIEM176-infected animals, and 2.5 × 106 (95% CI, 1.6 × 105–4 × 107) parasites/ml in TvMT1-infected ones. Anemia and clinical manifestations were more severe in the animals infected by TvMT1 strain than in those infected by TvLIEM176. In the proteomic analysis, a total of 29 proteins were identified, of which 14 exhibited significant differences in their expression levels between strains. Proteins with higher expression in TvLIEM176 were: alpha tubulin, beta tubulin, arginine kinase, glucose-regulated protein 78, paraflagellar protein 3, and T-complex protein 1 subunit theta. Proteins with higher expression in TvMT1 were: chaperonin HSP60, T-complex protein 1 subunit alpha, heat shock protein 70, pyruvate kinase, glycerol kinase, inosine-5'-monophosphate dehydrogenase, 73 kDa paraflagellar rod protein, and vacuolar ATP synthase. There was a difference in the virulence and pathogenicity between the T. vivax strains: TvLIEM176 showed high virulence and moderate pathogenicity, whereas TvMT1 showed low virulence and high pathogenicity. The proteins identified in this study are discussed for their potential involvement in strains’ virulence and pathogenicity, to be further defined as biomarkers of severity in T. vivax infections.
1. Introduction
Cattle trypanosomosis, a very important livestock disease, is caused by Trypanosoma congolense, T. brucei and T. vivax in Africa (Auty et al., 2015, Chamond et al., 2010). In Latin America, bovine trypanosomosis is caused by T. vivax and T. evansi, which are mechanically transmitted by blood-sucking insects (Desquesnes, 2004, Ramírez-Iglesias et al., 2017). This disease is characterized by fluctuating parasitemia, fever, anemia, lethargy, loss of weight, decrease in milk production, reproductive failures and, eventually, death, causing significative losses in the livestock industry. Epidemiological studies in Venezuela have shown serological prevalence values, determined by ELISA, between 20.8 to 33.1% for T. vivax (Suárez et al., 2009, Toro et al., 1980).
It has been proposed that the severity of the disease depends on parasite strain, endemicity and host species (Auty et al., 2015, Batista et al., 2007, Morrison et al., 2016). However, the discrete roles the host, the parasite, and their relationships at the molecular level play in the different clinical disease presentations are poorly understood. The exploration of host-parasite interactions could lead to the development of improved diagnostic tools, new drugs, and/or vaccine strategies.
Vaccine control of trypanosomosis is strongly limited by the complexity of the parasite’s antigenic repertoire. A further challenge is that knowledge about trypanosome antigenic variation comes primarily from the T. brucei model where variant surface glycoprotein (VSG) is highly immunogenic. T. vivax expression of the VSG repertoire differs from that of T. brucei, and this protein family is not the most immunodominant antigen in this species (Fleming et al., 2016, Greif et al., 2013, Jackson et al., 2012, Ramirez-Barrios et al., 2015). The identification of invariant trypanosome components rather than variant surface proteins as potential targets for limiting infection or infection-mediated disease has been the focus of some previous studies (Black and Mansfield, 2016, Grébaut et al., 2009); thus, characterization of pathogenic or virulent biomarkers is highly relevant.
The extracellular localization of these trypanosome species within their mammalian hosts results in extensive molecular interactions with this environment. Previous studies of T. vivax in different hosts have demonstrated that parasite and host genotypes have a major impact on infection progression (de Gee et al., 1979, 1981, 1982). Differences have been found in the susceptibility of inbred mouse strains to infections with T. vivax isolates of medium or low virulence; however, no resistant model has been identified (de Gee et al., 1982). Although mouse models have been useful to understand the course and disease progression in T. vivax-infected animals (Blom-Potar et al., 2010, Chamond et al., 2010), the use of natural hosts (ruminants) will result in more efficient development of relevant and effective tools to combat this infectious disease.
The outcome of host-parasite interaction is determined by the nature of the damage that results from the molecular battle. Disease occurs when the host sustains sufficient damage to perturb homeostasis. Historically, pathogenicity and virulence have been difficult to define. In this study, pathogenicity is defined as the ability of a microorganism to produce disease and/or mortality (Devera et al., 2003, Pirofski and Casadevall, 2012). Moreover, virulence is a relative term and, according to modern definitions, is the ability of a pathogen to multiply inside a susceptible host and to modulate the host response (Devera et al., 2003, Holzmuller et al., 2008). Therefore, this means that virulence is a dependent variable that is contingent on the availability of a susceptible host and should be studied within the context of the host-pathogen interaction (Casadevall and Pirofski, 2001).
The aims of this study were to evaluate the virulence and pathogenicity of two T. vivax strains in experimentally-infected sheep, and identify the differentially expressed proteins between them with a combination of 2-D differential in-gel electrophoresis (DIGE) and mass spectrometry (MS).
2. Materials and methods
2.1. Parasites
The strains used in the experiments, TvLIEM176 (a gift from Dr. Laura Moron and Dr. Glenda Moreno, Los Andes University) and TvMT1, were obtained from naturally-infected cattle from different locations in Venezuela: TvLIEM176 from Trujillo State (Western region) and TvMT1 from Monagas State (Eastern region) as previously described (Gómez-Piñeres et al., 2014). Diagnosis of infection was performed by microhematocrit analysis (Woo, 1969) and polymerase chain reaction (PCR) as described (Masake et al., 1997). Blood was collected in tubes containing 1 mg/ml of ethylenediaminetetracetic acid (EDTA), mixed with 10% glycerol in 20 mM phosphate buffered saline (PBS) pH 7.2 containing 1% glucose (PBSG) and frozen in liquid nitrogen (Gómez-Piñeres et al., 2009, Ndao et al., 2004).
2.2. Experimental infections
Nine adult Barbados Blackbelly sheep, eighteen months old, weighing 20 kg, and negative for T. vivax infection (determined by ELISA and PCR tests) were purchased from a local market. Tests for gastrointestinal parasites (fecal flotation test) and liver fluke (fecal sedimentation test) were performed at time of purchase. Three animals were positive for gastrointestinal nematodes. Ivermectin at a dose of 0.2 mg/kg was administered to all animals. Two weeks before and during experimental infection, all animals were maintained under veterinary supervision in order to monitor animal health and welfare at Centro Experimental de Producción Animal (CEPA, College of Veterinary Medicine, University of Zulia) in fly-proof pens. Three sheep were inoculated intravenously with each isolate (106 parasites/animal) and three were used as control. After 60-day infection period, the animals were treated with diminazene aceturate (3.5 mg/kg). Clinical and hematological tests were performed on a weekly base during 4 additional weeks in order to ensure that they were cured. Protocols used in this study were approved by the University of Zulia Scientific Committee (protocol CC-0497-12) according to the code for animal experimentation and based on Venezuelan law (MCT-FONACIT, 2002).
2.3. Determination of virulence and pathogenicity patterns
In order to determine the virulence pattern of each isolate, parasite presence in blood was determined by the microhematocrit centrifuge technique (Woo, 1969) and then, parasitemia was measured every three days during the complete infection period by the hemocytometer method (Valera et al., 2005). Every three days, pathogenicity of each isolate was evaluated by clinical and hematological evaluations in each animal as follows:
2.3.1. Animals were examined for clinical signs of trypanosomosis by collection and analysis of the following parameters: rectal temperature (ºC), heart and respiration rates, color of the mucous membranes, size of palpable lymph nodes, hydration status, body condition, response to external stimuli, posture, coat condition, presence of lacrimation, subcutaneous edema, and general attitude.
2.3.2. Blood samples were collected from the jugular vein using vacutainer® tubes with EDTA as anticoagulant. Packed cell volume (PCV) levels were measured as an estimation of anemia using capillary microhematocrit centrifugation, and hemoglobin was determined spectrophotometrically by the cyanmethemoglobin method (Jain, 1986). White blood cells (WBC) counts by the hemocytometer method and leukocyte profiles were collected as described (Jain, 1986).
2.4. Statistical analysis
Linear mixed effects models were built to estimate the effect of the treatment (inoculation with TvLIEM176 strain, inoculation with TvMT1 strain, non-infected control group), day (categorical with 21 levels: 0 to 60 dpi), and their interaction on the different outcome variables: PCV, hemoglobin, WBC, lymphocytes, neutrophils, monocytes, eosinophils, parasitemia, body temperature, heart rate, and respiratory rate, with sheep as the random intercept. In the models for WBC, lymphocytes, neutrophils, monocytes, and eosinophils, a random coefficient for day (deviation of the sheep-specific coefficient of day from the average coefficient of day) was included. Examples of the linear mixed models can be found in the supplemental material (supplementary statistical information). Parasitemia values were log10-transformed, and the control group (non-infected) was not included in this particular analysis since control sheep had no parasitemia during the study. REML was used as the estimation method, and Kenward-Roger method was used to calculate the degrees of freedom. Adjusted means are reported with 95% confidence intervals (95% CI). Pairwise comparisons at each day (corrected for multiple comparisons using Sidak’s method) as well as overall treatment effect (F test including both simple effects and interactions) were performed. Significance level was set a priori at 0.05. All statistical analyses were performed with Stata 15.1 software (StataCorp, USA).
2.5. Protein purification
An additional four sheep meeting the criteria in Section 2.2 “Experimental Infections” were purchased for acquisition of T. vivax for proteomic analysis. Two sheep were infected with each isolate; however, only one animal developed parasitemia with TvMT1 isolate, whereas the two were parasitized with TvLIEM176 isolate. The parasitized sheep and samples derived from them were henceforth identified with “TvMT1”, “LIEM 1” and “LIEM 2” labels, respectively. Trypanosomes were purified from infected animals when parasitemia reached values of 2 × 107 parasites/ml or higher, according to the protocol previously described (González et al., 2005). Subsequently, the parasite membranes were disrupted by 4 cycles of freezing in liquid nitrogen and thawing at room temperature, and the parasite homogenate was centrifuged to recover the soluble protein fraction (supernatant). Later, the samples were subjected to precipitation with 2-D Clean-Up Kit (GE Healthcare Life Sciences) according to the manufacturer’s protocol. Protein from a sample of pre-obtained Venezuelan T. evansi isolate, TeAp-ElFrio01 (Perrone et al., 2009, Sánchez et al., 2015), was also precipitated to be utilized in subsequent proteomic comparisons. The resulting pellet from each sample was lyophilized and stored at −20 °C.
2.6. Preparation of protein samples and DIGE labelling
In preparation for 2D-DIGE/LC-MS/MS, lyophilized protein samples were rehydrated in 50 μl of solubilization buffer (7 M urea, 2 M thiourea, 4% w/v CHAPS, 30 mM Tris, 0.5% Triton X-100) and cOmplete™ Protease Inhibitor Cocktail (Roche) overnight at room temperature. The total protein concentration of each sample was determined by Bradford assay (Bradford, 1976).
For 2D-DIGE separation, proteins were labeled according to the manufacturer’s instructions (GE Healthcare). For each sample (TvMT1, LIEM 1, LIEM 2, and T. evansi), 50 μg of protein was labelled with 400 pmol of Cy3 fluorochrome and, as a separate technical replicate, with Cy5 fluorochrome (CyDyesPM, GE Healthcare) for 30 min on ice in the dark. This dye swap labelling of technical replicates eliminated specific dye effects from the subsequent pairwise comparisons in four gels represented in Table S1. An internal standard, containing equal amounts of each protein extract, was labelled with Cy2 fluorescent dye, and used in all gel comparison experiments to normalize protein levels across gels and to control gel-to-gel variation (Alban et al., 2003).
2.7. Two-dimensional gel electrophoresis
The Cy2 standard, one sample labelled with Cy3 and another sample labelled with Cy5 were mixed (150 μg total protein) in a rehydration solution (7 M Urea, 2 M thiourea, 4% CHAPS, 0.5% Triton X-100, 12 μl/ml Destreak Reagent (GE Healthcare) and 1% immobilized pH gradient (IPG) buffer, pH 3–10). The proteins were separated in the first dimension within 24 cm immobilized pH gradient strips, pH 3–10, non-linear (NL) (GE Healthcare), which were passively rehydrated for 14 h. Isoelectric focusing was performed using an Ettan IPGphor system 3 (GE Healthcare) as follows: 3 h at 60 V, 4 h gradient to 1000 V, 4 h gradient to 8000 V and 8000 V constant to reach a total of 80,000 V h. After isoelectric focusing, strips were incubated in equilibration solution (6 M urea, 300 mM Tris pH 8.8, 0.2% SDS, 30% glycerol, 1% DTT) for 15 min, followed by a second 15 min incubation step with the same equilibration buffer in which DTT was replaced by 2.5% iodoacetamide. The equilibrated IPG strips were positioned on top of an SDS-polyacrylamide gel (12% acrylamide, resolution 10–100 kDa) and sealed with 1% agarose for second dimension separation. The electrophoresis was performed using an Ettan Dalt Twelve gel caster system (GE Healthcare) at 17 °C and 17 mA/gel.
2.8. Image analysis and statistics
The gels were scanned and visualized using a Typhoon 9400 (GE Healthcare) using appropriate wavelengths and filters for Cy2: 490 nm, Cy3: 550 nm, and Cy5: 650 nm. Spot detection and quantification were performed with Progenesis SameSpots v3.0 software (Nonlinear Dynamics Ltd.). Spot quantitation was normalized using a Cy2-labelled internal standard. The volume of each spot was calculated as the product of spot area and spot intensity. Differential expression analysis was performed by comparing matched spots volumes between the T. vivax isolates. Protein expression of T. evansi was used as an exclusion filter. To select relevant spots, the one-way ANOVA test (P-value ≤0.05 and a fold change ≥2) provided in the SameSpots software (Table S2) was used.
2.9. Protein gel excision and digestion
Gels were stained using Coomassie Brilliant Blue and the spots of interest were manually excised from each gel in a laminar flow hood. Proteins were in-gel digested with trypsin as previously described (Wilm et al., 1996).
2.10. Nano LC-MS/MS and protein identification
Peptide samples were dehydrated in a vacuum centrifuge and then solubilized in 5 μl of 0.1% formic acid/2% acetonitrile. Three (3) μl were analyzed online by nano-flow HPLC-nanoelectrospray ionization using an LTQ Orbitrap XL mass spectrometer (LTQ Orbitrap XL, Thermo Fisher Scientific, San Jose, CA) coupled with an Ultimate 3000 HPLC (Dionex). Desalting and pre-concentration of samples were performed on-line on a Pepmap® precolumn (0.3 mm x 10 mm). A gradient consisting of 0–40 % A in 30 min, 80 % B in 15 min (A = 0.1 % formic acid, 2 % acetonitrile in water; B = 0.1 % formic acid in acetonitrile) at 300 nl/min was used to elute peptides from the capillary (0.075 mm x 150 mm) reverse-phase column (Pepmap®, Dionex). LC-MS/MS experiments comprised cycles of 5 events: an MS1 scan with orbitrap mass analysis at 60,000 resolution followed by collision-induced dissociation (CID) of the five most abundant precursors. Fragment ions generated by CID were detected at the linear trap. Normalized collision energy of 35 eV and activation time of 30 ms were used for CID. All spectra were recorded under positive ion mode using the Xcalibur 2.0.7 software (Thermo Fisher Scientific) with the instrument operating in the information-dependent acquisition mode throughout the HPLC gradient. The mass scanning range was m/z 400–2000 and standard mass spectrometric conditions for all experiments were: spray voltage, 2.2 kV; no sheath and auxiliary gas flow; heated capillary temperature, 200 °C; capillary voltage, 40 V and tube lens, 120 V. For all full scan measurements with the Orbitrap detector a lock-mass ion from ambient air (m/z 445.120024) was used as an internal calibrant as described (Olsen et al., 2005).
All MS/MS spectra were searched against the Trypanosoma entries of either SwissProt or TrEMBL databases (http://www.uniprot.org/; v 2012_07) by using the Proteome Discover software v 1.3 (Thermo Fisher Scientific) and Mascot v 2.3 algorithm (http://www.matrixscience.com/) with trypsin enzyme specificity and one trypsin missed cleavage. Carbamidomethylation was set as fixed cysteine modification and oxidation was set as variable methionine modification for searches. A peptide mass tolerance of 5 ppm and a fragment mass tolerance of 0.5 Da were allowed for identification.
Management and validation of mass spectrometry data were carried out using Proteome Discoverer software v 1.3 (P < 0.01 for 2 peptides or more/protein). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD013266.
2.11. Bioinformatics analysis
Differentially expressed peptides between strains were utilized to search the T. vivax transcriptome database (http://bioinformatica.fcien.edu.uy/Tvivax/) for the amino acid sequence of each full-length T. vivax protein (Greif et al., 2013). This database was generated from the TvLIEM176 strain used in this study.
3. Results
3.1. T. vivax strains TvMT1 and TvLIEM176 show different virulence and pathogenicity patterns
The host-pathogen interactions in animal trypanosomosis must be analyzed from the parasite and the host perspectives; as host damage is induced by both the parasite and the host’s own immune response. Here, parasitemia levels as well as clinical and hematological observations associated with experimental infections of two T. vivax isolates in sheep were evaluated.
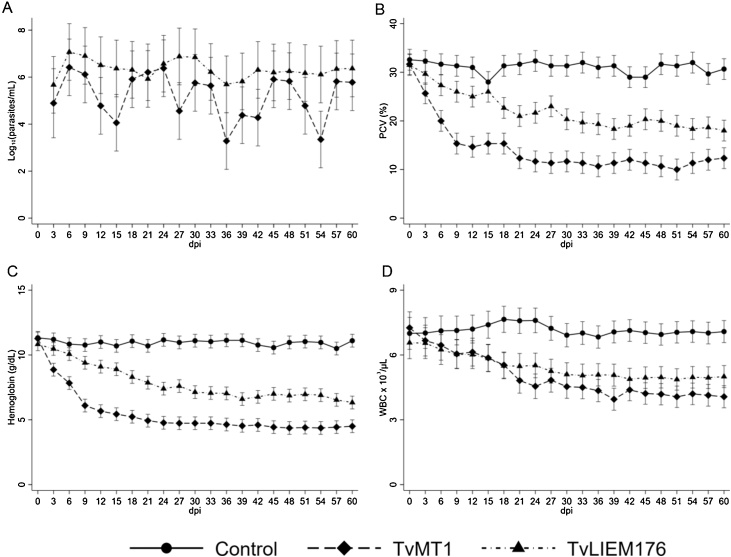
Three (all) TvLIEM176- and two TvMT1-infected animals developed parasitemia at 3 dpi, but the first peak in levels of blood parasitemia was observed at 6 dpi (Fig. S1, Table S3). The maximum average of parasitemia reached in the first peak (6 dpi) was 1.3 × 107 (95% CI, 7.9 × 105 – 2 × 108) parasites per ml of blood in TvLIEM176-infected animals and 2.5 × 106 (95% CI, 1.6 × 105 – 4 × 107) parasites per ml of blood in TvMT1-infected animals (Figs. 1A, S1). Then, parasitemia fluctuated for the rest of the trial period. Although parasitemia values were higher in TvLIEM176 infected animals than in TvMT1 ones during the infection period, statistical differences (P < 0.05) were observed only on 15 (P = 0.01), 27 (P = 0.009), 36 (P = 0.007), 42 (P = 0.022) and 54 (P = 0.002) dpi (Fig. 1A, Table S3).
Fig. 1.
Hematological analysis of non-infected and experimentally-infected sheep with Trypanosoma vivax strains TvMT1 and TvLIEM176. Adjusted mean values and 95% confidence intervals of parasitemia (log10 parasites/ml, panel A), PCV (%, panel B), hemoglobin (g/dl, panel C), and WBC counts (cells x 103/μl, panel D) during infection. Each group consisted of three sheep.
Anemia was important in the infected animals during the experiment. PCV and hemoglobin showed similar curves with a decrease in the values during the experimental period (Fig. 1B, 1C). The mean reduction of PCV and hemoglobin was higher in the animals infected with the TvMT1 strain (47.3% and 43.6%, respectively) than in the group infected with TvLIEM176 strain (25.3% and 25.4%, respectively). Both infected groups showed significant statistical differences in PCV values between them (P < 0.001) and when compared with the control group (P < 0.001) (Fig. 1B), except on 15 dpi. After 6 dpi, hemoglobin values were statistical different (P < 0.001) between both infected groups, being lower in TvMT1-infected animals (Fig. 1C). Taken together, these alterations indicate that TvMT1 isolate was capable of producing more severe anemia than TvLIEM176 isolate.
WBC (Fig. 1D) count was reduced in both infected groups (TvMT1 and TvLIEM176) and reached values very close to the normal lower limit for sheep. There were significant statistical differences in the values of both infected groups when compared with the control group from 15 dpi until the end of the experimental period (P < 0.001). There was a statistically significant difference between the WBC values of infected groups at 39 dpi (P = 0.021) only (Fig. 1D). The absolute lymphocytes count globally decreased in both infected groups compared with control, but TvMT1-infected animals exhibited peak lymphocyte levels between 3 and 18 dpi, whereas TvLIEM176-infected animals exhibited lymphocyte counts steadily decreasing from the beginning of the infection (Fig. 2A) until recovery at the very end of the infection. In both cases, after 18 dpi there were statistical differences between infected and control groups (P < 0.001).
Fig. 2.
Adjusted mean absolute values and 95% confidence intervals for blood concentrations of lymphocytes (cells x 103/μl, panel A), neutrophils (cells x 103/μl, panel B), monocytes (cells x 102/μl, panel C), and eosinophils (cells x 103/μl, panel D) of non-infected and experimentally-infected sheep with Trypanosoma vivax strains TvMT1 and TvLIEM176. Each group consisted of three sheep.
Neutrophil, monocyte and eosinophil counts varied during the experimental period, but they always stayed within normal ranges for sheep (Fig. 2B, 2C, 2D). Neutrophils had a slight but constant decrease during the experimental period in TvLIEM176-infected animals. In TvMT1-infected animals, neutrophils had a dramatic decrease until 9 dpi and then there was a slight recovery between 9 and 18 dpi, after which levels remained stable until the end of the study. Neutrophil counts were always lower in TvMT1- than in TvLIEM176- infected animals (Fig. 2B).
Monocyte counts were higher in TvLIEM176-infected animals compared with the TvMT1-infected and control groups, with comparable fluctuations during the experimental period. In TvMT1-infected animals, monocytes had a peak between 12 and 18 dpi and then, they had a similar profile than in the animals from the control group. Globally, monocytes counts were always greater in TvLIEM176 group than those in TvMT1 group (Fig. 2C).
Eosinophil levels in TvMT1-infected and control animals were similar, with low numbers of circulating eosinophils. Conversely, animals infected with TvLIEM176 had a significant increase of eosinophils between 6 and 36 dpi compared with control and TvMT1 groups, and eosinophil levels were higher in TvLIEM176- than in TvMT1- infected animals during the complete experimental period (Fig. 2D).
Broadly, TvLIEM176 strain induced a less severe anemia compared with TvMT1. This is similar to the relatively milder phenotype of the WBC analyses in composite for TvLIEM176: TvLIEM176 induced a lower decrease in WBC, mainly a lower decrease in neutrophils, higher decrease in lymphocytes, and increase in monocytes and eosinophils compared with TvMT1. The complete hematological data are presented in the Table S3, Figs. S1 and S2.
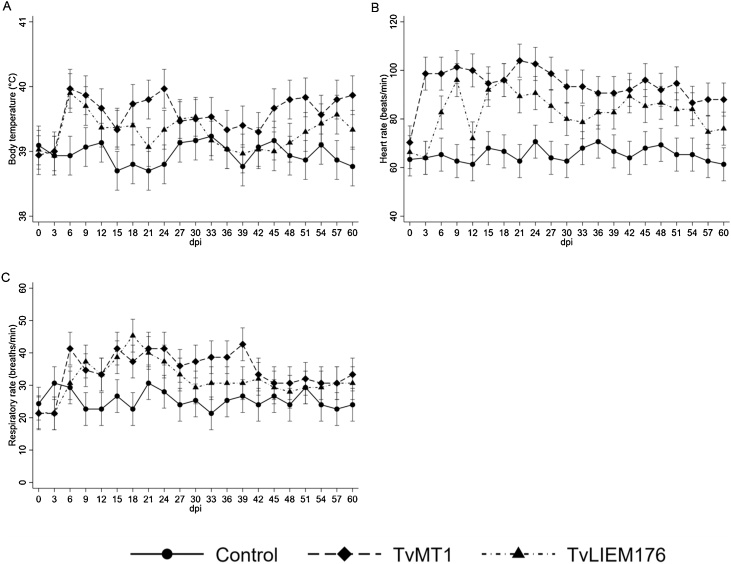
Vital signs (body temperature, heart rate, and respiratory rate) of sheep infected with both T. vivax isolates were recorded throughout the experiment (Fig. 3). The mean body temperatures of infected groups (39.6 °C [95% CI, 39.3 °C–39.9 °C], 39.3 °C [95% CI, 39.0 °C–39.6 °C]) for TvMT1 and TvLIEM176, respectively) were slightly higher than that of the control group (39.0 °C [95% CI, 38.7 °C–39.3 °C]). The body temperature of infected animals started rising from 6 dpi, coincident with the first peak of parasitemia, and then fluctuated throughout the study period (Fig. 3A). The highest mean body temperature recorded was 40.0 °C (95% CI, 39.7 °C–40.3 °C) from the TvMT1-infected animals at 6 and 24 dpi. Significant statistical differences were found between control non-infected group and the TvMT1 group from 6 to 24 dpi (P < 0.001) and on 39 (P = 0.012), 48 (P < 0.001), 51 (P < 0.001), 57 (P < 0.001), and 60 (P < 0.001) dpi. TvLIEM176-infected animals only had fever peaks on 6, 9 and 57 dpi (Fig. 3A).
Fig. 3.
Vital signs of non-infected and experimentally-infected sheep with Trypanosoma vivax strains TvMT1 and TvLIEM176. Adjusted mean values and 95% confidence intervals of body temperature (°C, panel A), heart rate (beats/min, panel B), and respiratory rate (breaths/min, panel C) during infection. Each group consisted of three sheep.
The mean baseline heart rate of the three groups of animals was similar at day 0 pre-inoculation (Control: 63.3 [95% CI, 56.5–70.1] beats/min, TvMT1-inoculated: 70.3 [95% CI, 63.5–77.1] beats/min, TvLIEM176-infected: 66.3 [95% CI, 59.5–73.1] beats/min). In the animals inoculated with TvMT1 isolate, the heart rate began to increase by day 3 following infection (98.7 [95% CI, 91.9–105.5] beats/min), reached the highest values (104 [95% CI, 97.2–110.8] beats/min) at 21 dpi and then had the tendency to keep stable around 90 beats/min during the rest of the study. TvLIEM176-infected animals showed a delayed and less marked increase of their heart rate from 6 dpi, and their average highest value (96 [95% CI, 89.2–102.8] beats/min at 9 and 18 dpi) was not as high as that in TvMT1-infected animals (Fig. 3B). The heart rate changes recorded in both infected groups were significantly higher (P <0.001 ) than those observed in the non-infected group. There were statistically significant heart rate differences on 3 (P < 0.001), 6 (P = 0.004), 12 (P < 0.001), 21–33 (P < 0.001), 57 (P = 0.023) and 60 (P = 0.047) dpi between both infected groups (Fig. 3B).
Respiratory rates followed a similar pattern (Fig. 3C). There was a fast increase of respiratory rate in TvMT1-infected animals from around 20 to 41 breaths/min during the first six days of infection, oscillating between 33 to 42 breaths/min until 39 dpi. Then, there was a decrease to about 30 breaths/min until the end of the study. In TvLIEM176-infected group, the increase in respiratory rate was slower, reaching 45 breaths/min at 18 dpi. A persistent decrease to around 29–30 breaths/min occurred from 30 dpi (Fig. 3C). Complete vital signs data are presented in the Table S3 and Fig. S3.
The clinical outcome of the disease was characterized by a variety of clinical signs. Initially, reduced feed intake and lethargy were observed. Later, facial edema, enlarged superficial lymph nodes, unilateral and bilateral lacrimation, pale or icteric membrane mucous, and weight loss were also observed. More severe clinical signs (icteric membrane mucous and harsh emaciation) were observed in the three TvMT1-infected animals.
3.2. T. vivax strains with different virulence and pathogenicity patterns exhibit differential expression of some proteins
After separating sample-specific labeled proteins by DIGE as described in Materials and Methods Sections 2.6 and 2.7, 754 spots were detected by SameSpots software in the DIGE gel images from T. vivax TvLIEM176 and TvMT1 strains. Spot intensity levels were quantitated and normalized using a Cy2-labeled internal standard. Seventeen protein spots which had at least a 2-fold higher intensity (ANOVA P < 0.05, Power >0.9) in T. vivax, compared with T. evansi, were initially identified. These 17 T. vivax-specific spots were then analyzed to identify those with at least a 2-fold difference in intensity (ANOVA P < 0.05, Power >0.9) between the T. vivax strains (Fig. 4). Six spots exhibited higher intensity in TvMT1 (758, 793, 1912, 1909, 1903, and 1906), and four in TvLIEM176 (1919, 1979, 1986, and 559). The rest of the spots (1552, 1099, 985, 602, 845, 1928, and 1969) were equally intense in both T. vivax strains. All 17 spots selected in the first screening (T. vivax-specific) were excised and analyzed by MS/MS. No spectra were recovered from one TvMT1 high-intensity spot (1909) but the spectra from the remaining 16 spots were analyzed: five more intense in TvMT1, four in TvLIEM176, and seven with similar intensity in both strains.
Fig. 4.
Two-dimensional synthetic image of differential in-gel electrophoresis gels displaying differentially expressed protein spots identified in the proteome of Trypanosoma vivax TvMT1 and TvLIEM176 strains.
Among these 16 spots, 29 peptides were consistent with proteins in the Uniprot database. These peptides were used to BLAST the T. vivax transcriptome database (Greif et al., 2013). Six proteins were identified from the spots displaying higher intensity in TvLIEM176, putatively annotated as alpha tubulin, beta tubulin, arginine kinase, glucose-regulated protein 78, paraflagellar protein 3, and T-complex protein 1 subunit theta. Eight proteins were identified from the spots displaying higher intensity in TvMT1, putatively annotated as heat shock protein 70 (HSP70), chaperonin HSP60, T-complex protein 1 subunit alpha, pyruvate kinase, glycerol kinase, inosine-5'-monophosphate dehydrogenase, 73 kDa paraflagellar rod protein, and vacuolar ATP synthase (Table 1). Proteins upregulated in TvLIEM176 strain are potentially associated with higher virulence but lower pathogenicity, and proteins upregulated in TvMT1 strain with higher pathogenicity but lower virulence. These proteins recovered from spots of strain-specific intensity were also classified according to their properties as vaccine candidates, potential drug targets and diagnosis candidates given the ontological assignment already described in the literature (Table 1).
Table 1.
Differentially expressed proteins identified in the proteome of two Trypanosoma vivax isolates of different virulence and pathogenicity, indicating the expression.
| Protein a | SNb | Accession Number or ID c | Species or Taxonomy d | Peptides/Unique peptides | Identification in T. vivax transcriptome e | Homology with other trypanosomes f | kDa/pI g | AA/Sc h | Molecular Function i | Biological Function j | Prop k | Reference l | TvLIEM176 m | TvMT1 m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpha tubulin | 1919 1979 1986 |
P04106 | Tbr | 9/9 | TvMiraNov_c43 |
T. vivax Y486: 100% T. b. rhodesiense: 99.8% T. b. gambiense: 99.8% |
49.8/5.06 | 455/84.7 | GTPase activity, GTP binding | Protein polymerization, GTP catabolic process | DT | (Lama et al., 2012, Nanavaty et al., 2016, Werbovetz, 2002, Yakovich et al., 2006) | +++ | + |
| VC | (Li et al., 2007, Plouffe and Belosevic, 2006) | |||||||||||||
| V | (Grébaut et al., 2009) | |||||||||||||
| Beta tubulin | 1919 1979 1986 |
A1XXJ2 | Te | 15/3 | TvMiraNov_rep_c5263 |
T. vivax Y486: 100% T. b. rhodesiense: 99.8% T. equiperdum: 99.8% |
49.6/4.87 | 442/132 | GTP binding, GTPase activity | GTP catabolic process, protein polymerization | DT | (Lama et al., 2012, Nanavaty et al., 2016, Werbovetz, 2002, Yakovich et al., 2006) | +++ | + |
| VC | (Li et al., 2007, Plouffe and Belosevic, 2006) | |||||||||||||
| V | (Grébaut et al., 2009) | |||||||||||||
| Arginine kinase | 1919 | O96507 | Tc | 4/4 | TvMiraNov_c1046 |
T. vivax Y486: 99.2% T. cruzi: 85.2% T. b. gambiense: 80.3% |
40.2/6.74 | 387/87 | Kinase activity, transferase activity | Metabolic process | V and P | (Grébaut et al., 2009) | +++ | + |
| DT | (Pereira et al., 2002) | |||||||||||||
| Glucose-regulated protein 78 | 1986 | D0A7U5 | Tbg | 2/2 | TvMiraNov_c10506 |
T. vivax Y486: 100% T. b. gambiense: 99.4% T. equiperdum: 99.4% |
65.2/5.52 | 157/296 | ATP binding | Protein folding | VC | (Jensen et al., 2001) | +++ | + |
| DC | (Krautz et al., 1998) | |||||||||||||
| T-complex protein 1 subunit theta | 1986 | D0A3R6 | Tbg | 2/1 | TvMiraNov_rep_c12168 |
T. vivax Y486: 94.8% T. congolense: 87.6% T. b. brucei: 87.6% |
58.1/5.53 | 244/27.3 | ATP binding, unfolded protein binding | Protein folding, cellular protein metabolic process | V | (Grébaut et al., 2009) | +++ | + |
| Par3 | 559 | O18658 | Tc | 6/5 | TvMiraNov_c274 |
T. vivax Y486: 43.8% T. b. gambiense: 41.1% T. equiperdum: 41.1% |
68.7/6.18 | 595/27.3 | ATP binding, unfolded protein binding | Protein folding, cellular protein metabolic process | V | (Grébaut et al., 2009) | +++ | + |
| VC | (Abdille et al., 2008, Clark et al., 2005, Morell et al., 2006, Saravia et al., 2005) | |||||||||||||
| Chaperonin HSP60 | 758 1912 |
D0A349 | Tbg | 6/6 | TvMiraNov_c5617 |
T. vivax Y486: 100% T. equiperdum: 94.7% T. b. gambiense: 94.7% |
59.5/5.44 | 471/42.4 | ATP binding | Protein refolding, cellular protein metabolic process | VC | (Radwanska et al., 2000a, Radwanska et al., 2000b) | + | +++ |
| T-complex protein 1 subunit alpha | 758 | D0AAR8 | Tbg | 3/2 | GCI4HUN02JWIRF |
T. vivax Y486: 100% T. b. brucei: 81.9% T. equiperdum: 81.9% |
59.4/5.92 | 160/38.9 | ATP binding, unfolded protein binding | Protein folding, cellular protein metabolic process, regulation of cell cycle | DT | (Liang and MacRae, 1997) | + | +++ |
| Heat shock protein 70 (HSP70) | 758 | A4GVT7 | Tr | 4/4 | TvMiraNov_c155 |
T. vivax Y486: 99.7% T. congolense: 92.6% T. cruzi: 92% |
71/5.81 | 668/50.1 | ATP binding | Stress response | DC | (Bossard et al., 2010) | + | +++ |
| VC | (Kumari et al., 2008) | |||||||||||||
| DT | (Evans et al., 2010) | |||||||||||||
| Pyruvate kinase 2 | 758 | P30616 | Tbb | 3/3 | TvMiraNov_c516 |
T. congolense: 88.6% T. theileri.: 88.2% T. b. gambiense: 86.3% |
54.6/7.39 | 499/34.3 | Magnesium ion binding, catalytic activity, pyruvate kinase activity, potassium ion binding | Glycolytic process | VC | (Shin et al., 2004, Yadav et al., 2017) | + | +++ |
| DC | (Pinto Torres et al., 2018, Shin et al., 2004, Yadav et al., 2017) | |||||||||||||
| DT | (Ernest et al., 1998, Worku et al., 2015) | |||||||||||||
| Putative glycerol kinase, glycosomal | 793 | G0U3C9 | Tv | 6/5 | TvMiraNov_c7270 |
T. vivax Y486: 100% T. b. brucei: 79.7% T. b. gambiense: 79.2% |
56.4/8.37 | 240/39.3 | Glycerol kinase activity, phosphotransferase activity | Carbohydrate metabolic process, glycerol-3-phosphate metabolic process | DT | (Ohashi-Suzuki et al., 2011) | + | +++ |
| Inosine-5'-monophosphate dehydrogenase | 793 | F9WQY2 | Tv | 2/1 | TvMiraNov_c8463 |
T. vivax Y486: 94.7% T. b. brucei: 79.9% T. b. gambiense: 79.9% |
55.6/8.5 | 374/26.2 | Catalytic activity, IMP dehydrogenase activity, adenyl nucleotide binding | Purine nucleotide biosynthetic process, oxidation-reduction process | DT | (Wilson et al., 1994) | + | +++ |
| 73 kDa paraflagellar rod protein | 1903 1906 |
C9ZLC2 | Tbg | 4/3 | TvMiraNov_c959 |
T. vivax Y486: 100% T. theileri: 93.2% T. cruzi: 92.3% |
59.6/6.11 | 221/37.4 | Protein binding | Calmodulin binding | VC | (Abdille et al., 2008, Clark et al., 2005, Morell et al., 2006, Saravia et al., 2005) | + | +++ |
| Vacuolar ATP synthase | 1906 | D0A941 | Tbg | 10/9 | GCI4HUN02HGO9U |
T. b. gambiense: 100% T. b. brucei: 100% T. vivax: 100% |
55.6/5.6 | 160/79 | ATP binding, hydrolase activity | ATP hydrolysis | V and P | (Grébaut et al., 2009) | + | +++ |
Name of the protein.
Spot number in the gel (Fig. 4).
Accession number of the protein according to UniProtKB database (https://uniprot.org).
Species: Tbr, Trypanosoma brucei rhodesiense; Te, Trypanosoma evansi; Tc, Trypanosoma cruzi; Tbg, Trypanosoma brucei gambiense; Tr, Trypanosoma rangeli, Tbb, Trypanosoma brucei brucei; Tv, Trypanosoma vivax.
Identification of the protein in the T. vivax transcriptome database (http://www.bioinformatica.fcien.edu.uy/Tvivax/).
Homology with the same protein from other trypanosomes, according to UniprotKB database (https://uniprot.org).
Theoretical Mr and pI, not determined for fragments of proteins.
Number of amino-acid residues, according to the T. vivax transcriptome database (http://www.bioinformatica.fcien.edu.uy/Tvivax/). Score (%).
Molecular function, according to UniProtKB (https://www.uniprot.org) and TriTrypdb (http://tritrypdb.org/tritrypdb/) databases.
Biological function, according to UniProtKB (https://www.uniprot.org) and TriTrypdb (http://tritrypdb.org/tritrypdb/) databases.
Molecular properties according to the literature: V, virulence; P, pathogenicity, VC, vaccine candidate; DT, drug target; DC, diagnostic candidate (antigenicity and immunogenicity).
Literature referring protein properties.
Expression level of the protein in each T. vivax isolate: +, low expression; +++, high expression, according the spot determined by SameSpots™ software.
Mass spectrometry results evidenced several protein chains for a given identification (TvMiraNov_c43: spots 1979, 1919, 1986; TvMiraNov_rep_c5263: spots 1979, 1919, 1986; TvMiraNov_c5617: spots 758, 1912; TvMiraNov_c959: spots 1903, 1906).
4. Discussion
Proteomic tools are useful for probing host-parasite crosstalk. Theoretically and empirically, both genotypic and phenotypic differences between trypanosomes strains are likely associated with the disease outcome. In this study, it was used a natural model of infection (sheep) to study the different patterns of virulence and pathogenicity of two T. vivax strains and to identify proteins whose potential for strain-specific differential expression could be influencers of clinical features of infection by that strain.
Differences in the severity and progression of trypanosome-caused diseases (virulence and pathogenicity, as defined in this study) have been described for infections of T. bruceigambiense (Holzmuller et al., 2008, Kaboré et al., 2018), T. bruceirhodesiense (Maclean et al., 2007, Muchiri et al., 2015), T. congolense(Bengaly et al., 2002a, Bengaly et al., 2002b, Grébaut et al., 2009, Masumu et al., 2006), T. evansi (Perrone et al., 2018, Verdillo et al., 2012) and T. cruzi (Andrade et al., 1985, Belew et al., 2017, Henrique et al., 2016, Lauria-Pires and Teixeira, 1996). Similarly, differences in virulence and pathogenicity have been recognized between East and West African strains of T. vivax, the West African strains being generally more pathogenic to cattle (Gardiner, 1989). However, Latin American T. vivax infections are considered generally chronic and mild, with rare outbreaks of severe disease (Osório et al., 2008). This is the first study comparing the different virulence and pathogenicity patterns of two Venezuelan T. vivax strains isolated in animals. It was found that they exhibit differences in the severity of their disease profiles and differences in their expressed proteomes.
The clinical outcome in sheep infected with both strains was a three-phase disease pattern. The prepatent period between the parasite inoculation and the appearance of trypanosomes in blood was three days, and the animals remained asymptomatic during this time. Then, an acute period was observed, characterized by high, persistent, and fluctuating parasitemia, recurrent fever, a severe decrease of PCV, and several pathophysiological and clinical signs. Finally, animals developed sub-acute disease with lower and fluctuating parasitemia. In spite of gradual cessation of disease symptoms, PCV values remained low during the sub-acute phase. The same disease pattern in sheep was observed using other T. vivax isolates (Anosa and Isoun, 1980, Maikaje et al., 1991, Valera et al., 2005).
However, substantial strain-specific differences in parasitemia, clinical signs, and hematological parameters in T. vivax infections were observed. Animals infected with TvMT1 strain showed lower values of parasitemia when compared with TvLIEM176-infected animals, whereas the induced clinical signs were more severe in TvMT1 infections than in TvLIEM176 ones. Similarly, the consistent higher parasitemia observed with the TvLIEM176 strain was associated with a lower magnitude of global decrease in WBC. Specifically, in TvLIEM176-infected sheep, neutrophil decrease was less severe, yet lymphocyte decrease was more severe, and levels of monocytes and eosinophils were higher than in those in TvMT1 infected sheep. Such differences in immune cell populations between animals infected with each strain likely reflect some divergence in modulation of the host immune response to these strains. Such strain-specific effects have been well detailed in bovine trypanosomosis caused by T. congolense(Taylor, 1998, Taylor and Mertens, 1999) and to a lesser extent in mouse infections with T. vivax (Blom-Potar et al., 2010, Morrison et al., 2010).
Anemia became marked particularly in animals infected with the TvMT1 strain as the disease progressed. PCV decrease, and the consequent anemia, is the hallmark of T. vivax pathogenesis and is used as one of the main disease indicators (Osório et al., 2008). The mechanisms that generate anemia in trypanosomosis have not been totally elucidated and the pathogenesis of this feature is extremely complex and multifactorial. A number of factors have been incriminated in the etiology of anemia and these include hemolysis as a result of immunological factors and trypanosome products, hemodilution and impaired erythropoiesis (Katunguka-Rwakishaya et al., 1992). In infections with T. vivax, direct alteration of the red blood cells surface is expected to be an important mechanism in anemia pathogenesis due to the secretion of sialidases by these parasites, leading to their phagocytosis (Guegan et al., 2013). Overall, data collected from sheep infections demonstrated that TvLIEM176 strain had high virulence and moderate pathogenicity and TvMT1 strain had low virulence and high pathogenicity.
These striking differences led to the hypothesis that these two strains also differentially express their proteome when replicating in sheep. The spots in DIGE analysis that were particularly abundant in the T. vivax strains relative to their intensity in T. evansi, a different trypanosome species, were identified. However, we focused only on the differentially expressed proteins between both T. vivax strains. MS/MS spectra of these spots resulted in the identification of 29 proteins, of which 14 were found within spots that were differentially expressed between the high virulence/moderate pathogenicity and the low virulence/high pathogenicity T. vivax strains. Trypanosomes have acquired efficient immune evasion mechanisms to undermine protective host immune response and survive in the host’s extracellular environment (Stijlemans et al., 2017). Factors produced by the parasites would be necessary for this, and differences in expression of these factors could play a role in disease pathogenicity (Siqueira-Neto et al., 2018, Stijlemans et al., 2017, Taylor, 1998).
Proteins from DIGE spots that are more intense in the more virulent strain (TvLIEM176) were the tubulins (alpha and beta chains), which are a fundamental component of microtubules. The highly conserved eukaryotic tubulins are vital to cell division, motility, intracellular transport, and signal transduction (Lama et al., 2012). Despite tubulin sequence conservation (Werbovetz et al., 2003), there are differences in orthologue susceptibility to antimitotic agents, likely reflecting structural differences among species (Bobba et al., 2017, Lama et al., 2012, Werbovetz et al., 2003). The stable biological characteristics of trypanosome tubulin make it an ideal potential drug target and also a prospective vaccine candidate (Kurup and Tewari, 2012, Lama et al., 2012, Li et al., 2007, Nanavaty et al., 2016, Werbovetz, 2002). As tubulin peptides were identified in multiple spots, proteoforms varying in molecular weight and isoelectric point are likely, possibly corresponding to different maturation states and/or post-translational modifications. Different tubulin proteoforms might vary in functionality (Sasse and Gull, 1988), affecting progression through the trypanosome cell cycle, and thus virulence. Moreover, different secreted proteoforms of tubulin alpha and beta chains of T. congolensehave been reported as virulence factors (Grébaut et al., 2009). The importance of microtubules in intracellular transport (Montalvão et al., 2018) could also explain the likely higher expression of tubulin in the TvLIEM176 strain.
Another family of proteins related to parasite motility are the paraflagellar rod (PFR) proteins. Here, it was found a putative 73 kDa paraflagellar rod protein in DIGE spots of higher intensity in the strain exhibiting higher pathogenicity and a paraflagellar protein 3 homologue in higher intensity spots of the more virulent strain. The PFR, a unique attribute of kinetoplastids, is essential for motility and cell viability (Koyfman et al., 2011, Portman and Gull, 2010, Ralston and Hill, 2008). PFR proteins are considered potential drug targets and excellent vaccines candidates due to their high sequence conservation between kinetoplastid parasites and lack of human and livestock animal near orthologues (Abdille et al., 2008, Clark et al., 2005, Morell et al., 2006). Their potential as vaccine candidates has been demonstrated by immunization of hamsters with Leishmania mexicana PFR2 which resulted in delayed appearance of cutaneous lesions and significant reductions in lesion size (Saravia et al., 2005).
Two putative heat shock protein homologues were identified in DIGE spots that were more intense in the more pathogenic strain (TvMT1): HSP60 and HSP70. Conversely, the putative glucose-regulated protein 78 (GRP78), a member of the HSP70 family localized in endoplasmic reticulum, was found in the spots of higher intensity in the less pathogenic but more virulent strain (TvLIEM176). The heat shock proteins (HSPs) influence folding, assembly, intracellular localization, secretion, regulation and degradation of other proteins (Folgueira and Requena, 2007). In trypanosomatids, links between HSP70 and development and survival of parasites during temperature changes have been established (Folgueira and Requena, 2007, Louw et al., 2010). Also, HSPs are highly immunogenic and although they are very similar to their human and animal homologs, they possess many determinants that induce strong humoral and cellular immune responses during infection (Bossard et al., 2010, Krautz et al., 1998). Given their complex roles in disease presentation though, it would be difficult to define them precisely as drug targets (Evans et al., 2010).
Another differentially expressed protein in this study was the T. vivax homologue of the glycolytic enzyme pyruvate kinase (PK), which was more expressed by the TvMT1 strain. PK activity is relatively high in the T. brucei cytosol and is crucial for carbohydrate and energy metabolism (Ernest et al., 1998); therefore, it has been considered as a potential drug target. Pharmacological inhibition or RNAi silencing of this enzyme causes ATP depletion, cell growth arrest, and parasite death (Albert et al., 2005, Coustou et al., 2003, Worku et al., 2015). Additionally, the antigenic properties of PK in T. evansi and T. congolenseare known (Pinto Torres et al., 2018, Yadav et al., 2017), and PK has been identified as a diagnostic biomarker in T. congolense (Pinto Torres et al., 2018).
This study highlights the variation in protein expression between T. vivax strains of different geographical origins. The strains compared here were from very distant Venezuelan locations and environments. It is also showed differences in disease outcomes of these strains. It is possible but not proven that the molecular plasticity related to the differences in these two T. vivax strains is responsible for their differences in virulence and pathogenicity. For this study, the two available Venezuelan strains with the highest known pathogenicity and virulence were selected, but it might be possible to find additional differences in protein expression with strains from other regions. It is also important to consider host variability in susceptibility to infection, which was not considered here. Nevertheless, these results provide circumstantial evidence for varying virulence and pathogenicity patterns influenced by parasite genetic diversity or molecular plasticity.
Further studies are needed to establish a causal relationship between disease outcome and protein expression in T. vivax infections. Finally, additional studies are to validate the biological properties of the proteins identified here in order to consider their use in prevention, identification, or treatment of South American animal trypanosomosis caused by T. vivax.
Conflict of interest
The authors do not have any conflicts of interest to declare.
All animal infections were done in compliance with Venezuelan animal welfare regulations. The animal housing conditions and protocols used in the present research were previously approved by the University of Zulia Scientific Committee under the number CC-0497-12, according to Code of Bioethics and Biosecurity of the Venezuelan Council of Scientific and Technological Research (CONICIT), which includes appropriate procedures to minimize pain and animal suffering.
CRediT authorship contribution statement
Roger Ramirez-Barrios: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition. Armando Reyna-Bello: Conceptualization, Methodology, Validation, Investigation, Resources, Writing - original draft, Supervision. Omaira Parra: Methodology, Investigation. Robert Valeris: Methodology, Validation, Formal analysis, Investigation, Writing - review & editing. Lucinda Tavares-Marques: Investigation. Jean-Paul Brizard: Methodology, Investigation. Edith Demettre: Investigation. Martial Seveno: Investigation, Writing - review & editing. Alvaro Martinez-Moreno: Methodology, Writing - original draft, Writing - review & editing, Supervision. Philippe Holzmuller: Conceptualization, Methodology, Investigation, Resources, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgements
This research was supported by grants from ECOS NORD (Project Number V09A01) and from the Consejo de Desarrollo Cientifico, Humanistico y Tecnologico (CONDES) of the University of Zulia. Mass spectrometry experiments were carried out using facilities of the Functional Proteomics Platform of Montpellier. We are very thankful to Dr. Sara L. Zimmer for critically reading the manuscript.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:10.1016.vpoa.2019.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Abdille M.H., Li S.Y., Ding J., Suo X. Trypanosoma evansi: paraflagellar rod protein 1 and 2 are similar but lack common B cell epitopes. Exp. Parasitol. 2008;120:411–416. doi: 10.1016/j.exppara.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Alban A., David S.O., Bjorkesten L., Andersson C., Sloge E., Lewis S., Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- Albert M.A., Haanstra J.R., Hannaert V., Van Roy J., Opperdoes F.R., Bakker B.M., Michels P.A.M. Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei. J. Biol. Chem. 2005;280:28306–28315. doi: 10.1074/jbc.M502403200. [DOI] [PubMed] [Google Scholar]
- Andrade S.G., Andrade V., Brodskyn C., Magalhães J.B., Netto M.B. Immunological response of Swiss mice to infection with three different strains of Trypanosoma cruzi. Ann. Trop. Med. Parasitol. 1985;79:397–407. doi: 10.1080/00034983.1985.11811938. [DOI] [PubMed] [Google Scholar]
- Anosa V.O., Isoun T.T. Haematological studies on Trypanosoma vivax infection of goats and intact and splenectomized sheep. J. Comp. Pathol. 1980;90:155–168. doi: 10.1016/0021-9975(80)90036-5. [DOI] [PubMed] [Google Scholar]
- Auty H., Torr S.J., Michoel T., Jayaraman S., Morrison L.J. Cattle trypanosomosis: the diversity of trypanosomes and implications for disease epidemiology and control. Rev. Sci. Tech. 2015;34:587–598. doi: 10.20506/rst.34.2.2382. [DOI] [PubMed] [Google Scholar]
- Batista J.S., Riet-Correa F., Teixeira M.M.G., Madruga C.R., Simões S.D.V., Maia T.F. Trypanosomiasis by Trypanosoma vivax in cattle in the Brazilian semiarid: description of an outbreak and lesions in the nervous system. Vet. Parasitol. 2007;143:174–181. doi: 10.1016/j.vetpar.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Belew A.T., Junqueira C., Rodrigues-Luiz G.F., Valente B.M., Oliveira A.E.R., Polidoro R.B., Zuccherato L.W., Bartholomeu D.C., Schenkman S., Gazzinelli R.T., Burleigh B.A., El-Sayed N.M., Teixeira S.M.R. Comparative transcriptome profiling of virulent and non-virulent Trypanosoma cruzi underlines the role of surface proteins during infection. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengaly Z., Sidibe I., Boly H., Sawadogo L., Desquesnes M. Comparative pathogenicity of three genetically distinct Trypanosoma congolense-types in inbred Balb/c mice. Vet. Parasitol. 2002;105:111–118. doi: 10.1016/S0304-4017(01)00609-4. [DOI] [PubMed] [Google Scholar]
- Bengaly Z., Sidibe I., Ganaba R., Desquesnes M., Boly H., Sawadogo L. Comparative pathogenicity of three genetically distinct types of Trypanosoma congolense in cattle: clinical observations and haematological changes. Vet. Parasitol. 2002;108:1–19. doi: 10.1016/S0304-4017(02)00164-4. [DOI] [PubMed] [Google Scholar]
- Black S.J., Mansfield J.M. Prospects for vaccination against pathogenic African trypanosomes. Parasite Immunol. 2016;38:735–743. doi: 10.1111/pim.12387. [DOI] [PubMed] [Google Scholar]
- Blom-Potar M.C., Chamond N., Cosson A., Jouvion G., Droin-Bergère S., Huerre M., Minoprio P. Trypanosoma vivax infections: pushing ahead with mouse models for the study of nagana. II. Immunobiological dysfunctions. PLoS Negl. Trop. Dis. 2010;4:e793. doi: 10.1371/journal.pntd.0000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobba V., Nanavaty V., Idippily N.D., Zhao A., Li B., Su B. Synthesis and biological evaluation of selective tubulin inhibitors as anti-trypanosomal agents. Bioorg. Med. Chem. 2017;25:3215–3222. doi: 10.1016/J.BMC.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard G., Boulange A., Holzmuller P., Thévenon S., Patrel D., Authie E. Serodiagnosis of bovine trypanosomosis based on HSP70/BiP inhibition ELISA. Vet. Parasitol. 2010;173:39–47. doi: 10.1016/j.vetpar.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L. Host‐Pathogen interactions: The attributes of virulence. J. Infect. Dis. 2001;184:337–344. doi: 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- Chamond N., Cosson A., Blom-Potar M.C., Jouvion G., D’Archivio S., Medina M., Droin-Bergère S., Huerre M., Goyard S., Minoprio P. Trypanosoma vivax infections: pushing ahead with mouse models for the study of nagana. I. Parasitological, hematological pathological parameters. PLoS Negl. Trop. Dis. 2010;4:e792. doi: 10.1371/journal.pntd.0000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.K., Kovtunovych G., Kandlikar S., Lal S., Stryker G.A. Cloning and expression analysis of two novel paraflagellar rod domain genes found in Trypanosoma cruzi. Parasitol. Res. 2005;96:312–320. doi: 10.1007/s00436-005-1370-2. [DOI] [PubMed] [Google Scholar]
- Coustou V., Besteiro S., Biran M., Diolez P., Bouchaud V., Voisin P., Michels P.A.M., Canioni P., Baltz T., Bringaud F. ATP generation in the Trypanosoma brucei procyclic form: cytosolic substrate level is essential, but not oxidative phosphorylation. J. Biol. Chem. 2003;278:49625–49635. doi: 10.1074/jbc.M307872200. [DOI] [PubMed] [Google Scholar]
- de Gee A.L.W., Shah S.D., Doyle J.J. Trypanosoma vivax: sequence of antigenic variants in mice and goats. Exp. Parasitol. 1979;48:352–358. doi: 10.1016/0014-4894(79)90119-X. [DOI] [PubMed] [Google Scholar]
- de Gee A.L., Shah S.D., Doyle J.J. Trypanosoma vivax: host influence on appearance of variable antigen types. Exp. Parasitol. 1981;51:392–399. doi: 10.1016/0014-4894(81)90126-0. [DOI] [PubMed] [Google Scholar]
- de Gee A.L.W., Shah S.D., Doyle J.J. Trypanosoma vivax: courses of infection with three stabilates in inbred mouse strains. Exp. Parasitol. 1982;54:33–39. doi: 10.1016/0014-4894(82)90107-2. [DOI] [PubMed] [Google Scholar]
- Desquesnes M. OIE; Paris: 2004. Livestock Trypanosomoses and Their Vectors in Latin America. [Google Scholar]
- Devera R., Fernandes O., Coura J.R. Should Trypanosoma cruzi be called “cruzi” complex? A review of the parasite diversity and the potential of selecting population after in vitro culturing and mice infection. Mem. Inst. Oswaldo Cruz. 2003;98:1–12. doi: 10.1590/S0074-02762003000100001. [DOI] [PubMed] [Google Scholar]
- Ernest I., Callens M., Uttaro A.D., Chevalier N., Opperdoes F.R., Muirhead H., Michels P.A.M. Pyruvate kinase of Trypanosoma brucei: overexpression, purification, and functional characterization of wild-type and mutated enzyme. Protein Expr. Purif. 1998;13:373–382. doi: 10.1006/prep.1998.0918. [DOI] [PubMed] [Google Scholar]
- Evans C.G., Chang L., Gestwicki J.E. Heat shock protein 70 (Hsp70) as an emerging drug target. J. Med. Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J.R., Sastry L., Wall S.J., Sullivan L., Ferguson M.A.J. Proteomic identification of immunodiagnostic antigens for Trypanosoma vivax infections in cattle and generation of a proof-of-concept lateral flow test diagnostic device. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueira C., Requena J.M. A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol. Rev. 2007;31:359–377. doi: 10.1111/j.1574-6976.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Gardiner P.R. Recent studies of the biology of Trypanosoma vivax. Adv. Parasitol. 1989;28:229–317. doi: 10.1016/S0065-308X(08)60334-6. [DOI] [PubMed] [Google Scholar]
- Gómez-Piñeres E., Tavares-Marques L., Reyna-Bello A. Tiempo de supervivencia in vivo y criopreservación de Trypanosoma vivax. Rev. Cientif. FCV-LUZ. 2009;19:225–229. [Google Scholar]
- Gómez-Piñeres E., Boada-Sucre A., Bretaña A., Contreras-Bretaña M., García F., Reyna-Bello A. Morfometría comparativa de cinco aislados venezolanos de Trypanosoma vivax. Rev. Fac. Ciencias Vet. UCV. 2014;55:25–33. [Google Scholar]
- González L.E.E., García J.A.A., Núñez C., Perrone T.M.M., González-Baradat B., Gonzatti M.I.I., Reyna-Bello A. Trypanosoma vivax: A novel method for purfication from experimentally infected sheep blood. Exp. Parasitol. 2005;111:126–129. doi: 10.1016/j.exppara.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Grébaut P., Chuchana P., Brizard J.P., Demettre E., Seveno M., Bossard G., Jouin P., Vincendeau P., Bengaly Z., Boulangé A., Cuny G., Holzmuller P. Identification of total and differentially expressed excreted-secreted proteins from Trypanosoma congolense strains exhibiting different virulence and pathogenicity. Int. J. Parasitol. 2009;39:1137–1150. doi: 10.1016/j.ijpara.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Greif G., Ponce de Leon M., Lamolle G., Rodriguez M., Piñeyro D., Tavares-Marques L.M., Reyna-Bello A., Robello C., Alvarez-Valin F. Transcriptome analysis of the bloodstream stage from the parasite Trypanosoma vivax. BMC Genomics. 2013;14:149. doi: 10.1186/1471-2164-14-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guegan F., Plazolles N., Baltz T., Coustou V. Erythrophagocytosis of desialylated red blood cells is responsible for anaemia during Trypanosomavivax infection. Cell. Microbiol. 2013;15:1285–1303. doi: 10.1111/cmi.12123. [DOI] [PubMed] [Google Scholar]
- Henrique P.M., Marques T., da Silva M.V., Nascentes G.A.N., de Oliveira C.F., Rodrigues V., Gómez-Hernández C., Norris K.A., Ramirez L.E., Meira W.S.F. Correlation between the virulence of T.cruzi strains, complement regulatory protein expression levels, and the ability to elicit lytic antibody production. Exp. Parasitol. 2016;170:66–72. doi: 10.1016/j.exppara.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Holzmuller P., Biron D.G., Courtois P., Koffi M., Bras-Gonçalves R., Daulouède S., Solano P., Cuny G., Vincendeau P., Jamonneau V. Virulence and pathogenicity patterns of Trypanosoma brucei gambiense field isolates in experimentally infected mouse: differences in host immune response modulation by secretome and proteomics. Microbes Infect. 2008;10:79–86. doi: 10.1016/j.micinf.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Jackson A.P., Berry A., Aslett M., Allison H.C., Burton P., Vavrova-Anderson J., Brown R., Browne H., Corton N., Hauser H., Gamble J., Gilderthorp R., Marcello L., McQuillan J., Otto T.D., Quail M.A., Sanders M.J., van Tonder A., Ginger M.L., Field M.C., Barry J.D., Hertz-Fowler C., Berriman M. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc. Natl. Acad. Sci. 2012;109:3416–3421. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N.C. 4th ed. Lea & Febiger, Philadelphia; 1986. Schalm’s Veterinary Hematology. [Google Scholar]
- Jensen A.T., Curtis J., Montgomery J., Handman E., Theander T.G. Molecular and immunological characterisation of the glucose regulated protein 78 of Leishmania donovani. Biochim. Biophys. Acta. 2001;1549:73–87. doi: 10.1016/S0167-4838(01)00240-0. [DOI] [PubMed] [Google Scholar]
- Kaboré J., Camara O., Koffi M., Sanou D., Ilboudo H., Sakandé H., Camara M., De Meeûs T., Ravel S., Belem A.M.G., MacLeod A., Bucheton B., Jamonneau V., Thévenon S. Differences in pathogenicity and virulence of Trypanosoma brucei gambiense field isolates in experimentally infected Balb/C mice. Infect. Genet. Evol. 2018;63:269–276. doi: 10.1016/j.meegid.2018.05.018. [DOI] [PubMed] [Google Scholar]
- Katunguka-Rwakishaya E., Murray M., Holmes P.H. The pathophysiology of ovine trypanosomosis: haemotological and blood biochemical changes. Vet. Parasitol. 1992;45:17–32. doi: 10.1016/0304-4017(92)90024-4. [DOI] [PubMed] [Google Scholar]
- Koyfman A.Y., Schmid M.F., Gheiratmand L., Fu C.J., Khant H.A., Huang D., He C.Y., Chiu W. Structure of Trypanosoma bruceiflagellum accounts for its bihelical motion. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11105–11108. doi: 10.1073/pnas.1103634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautz G.M., Krettli A.U., Godsel L.M., Engman D.M., Peterson J.D. Human antibody responses to Trypanosoma cruzi 70-kD heat-shock proteins. Am. J. Trop. Med. Hyg. 1998;58:137–143. doi: 10.4269/ajtmh.1998.58.137. [DOI] [PubMed] [Google Scholar]
- Kumari S., Samant M., Misra P., Khare P., Sisodia B., Shasany A.K., Dube A. Th1-stimulatory polyproteins of soluble Leishmania donovani promastigotes ranging from 89.9 to 97.1 kDa offers long-lasting protection against experimental visceral leishmaniasis. Vaccine. 2008;26:5700–5711. doi: 10.1016/j.vaccine.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Kurup S.P., Tewari A.K. Induction of protective immune response in mice by a DNA vaccine encoding Trypanosoma evansi beta tubulin gene. Vet. Parasitol. 2012;187:9–16. doi: 10.1016/j.vetpar.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Lama R., Sandhu R., Zhong B., Li B., Su B. Identification of selective tubulin inhibitors as potential anti-trypanosomal agents. Bioorg. Med. Chem. Lett. 2012;22:5508–5516. doi: 10.1016/j.bmcl.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria-Pires L., Teixeira A.R.L. Virulence and pathogenicity associated with diversity of Trypanosoma cruzi stocks and clones derived from chagas’ disease patients. Am. J. Trop. Med. Hyg. 1996;55:304–310. doi: 10.4269/ajtmh.1996.55.304. [DOI] [PubMed] [Google Scholar]
- Li S.Q., Fung M.C., Reid S.A., Inoue N., Lun Z.R. Immunization with recombinant beta-tubulin from Trypanosomaevansi induced protection against T.evansi, T.equiperdum and T.b. brucei infection in mice. Parasite Immunol. 2007;29:191–199. doi: 10.1111/j.1365-3024.2006.00933.x. [DOI] [PubMed] [Google Scholar]
- Liang P., MacRae T.H. Molecular chaperones and the cytoskeleton. J. Cell. Sci. 1997;110:1431–1440. doi: 10.1242/jcs.110.13.1431. [DOI] [PubMed] [Google Scholar]
- Louw C.A., Ludewig M.H., Mayer J., Blatch G.L. The Hsp70 chaperones of the tritryps are characterized by unusual features and novel members. Parasitol. Int. 2010;59:497–505. doi: 10.1016/j.parint.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Maclean L., Odiit M., Macleod A., Morrison L., Sweeney L., Cooper A., Kennedy P.G.E., Sternberg J.M. Spatially and genetically distinct African trypanosome virulence variants defined by host interferon-gamma response. J. Infect. Dis. 2007;196:1620–1628. doi: 10.1086/522011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maikaje D.B.B., Sannusi A., Kyewalabye E.K.K., Saror D.I.I. The course of experimental Trypanosoma vivax infection in Uda sheep. Vet. Parasitol. 1991;38:267–275. doi: 10.1016/0304-4017(91)90139-M. [DOI] [PubMed] [Google Scholar]
- Masake R.A., Majiwa P.A.O., Moloo S.K., Makau J.M., Njuguna J.T., Maina M., Kabata J., Ole-Moiyoi O.K., Nantulya V.M. Sensitive and specific detection of Trypanosoma vivax using the polymerase chain reaction. Exp. Parasitol. 1997;85:193–205. doi: 10.1006/expr.1996.4124. [DOI] [PubMed] [Google Scholar]
- Masumu J., Marcotty T., Geysen D., Geerts S., Vercruysse J., Dorny P., Van den Bossche P. Comparison of the virulence of Trypanosoma congolense strains isolated from cattle in a trypanosomiasis endemic area of eastern Zambia. Int. J. Parasitol. 2006;36:497–501. doi: 10.1016/j.ijpara.2006.01.003. [DOI] [PubMed] [Google Scholar]
- MCT-FONACIT . 2002. Código de Bioética y Bioseguridad del Consejo Nacional de Investigaciones Científicas y Tecnológicas (Conicit) [Google Scholar]
- Montalvão F., Nascimento D.O., Nunes M.P., Koeller C.M., Morrot A., Lery L.M.S., Bisch P.M., Teixeira S.M.R., Vasconcellos R., Freire-de-Lima L., Lopes M.F., Heise N., DosReis G.A., Freire-de-Lima C.G. Antibody repertoires identify β-tubulin as a host protective parasite antigen in mice infected with Trypanosoma cruzi. Front. Immunol. 2018;9:671. doi: 10.3389/fimmu.2018.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M., Thomas M.C., Caballero T., Alonso C., López M.C. The genetic immunization with paraflagellar rod protein-2 fused to the HSP70 confers protection against late Trypanosoma cruziinfection. Vaccine. 2006;24:7046–7055. doi: 10.1016/J.VACCINE.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Morrison L.J., McLellan S., Sweeney L., Chan C.N., MacLeod A., Tait A., Turner C.M.R. Role for parasite genetic diversity in differential host responses to Trypanosoma bruceiinfection. Infect. Immun. 2010;78:1096–1108. doi: 10.1128/IAI.00943-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison L.J., Vezza L., Rowan T., Hope J.C. Animal African trypanosomiasis: time to increase focus on clinically relevant parasite and host species. Trends Parasitol. 2016;32:599–607. doi: 10.1016/j.pt.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Muchiri M.W., Ndung’u K., Kibugu J.K., Thuita J.K., Gitonga P.K., Ngae G.N., Mdachi R.E., Kagira J.M. Comparative pathogenicity of Trypanosoma brucei rhodesiense strains in swiss white mice and mastomys natalensis rats. Acta Trop. 2015;150:23–28. doi: 10.1016/J.ACTATROPICA.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Nanavaty V., Lama R., Sandhu R., Zhong B., Kulman D., Bobba V. Orally active and selective tubulin inhibitors as anti-trypanosome agents. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndao M., Magnus E., Büscher P., Geerts S. Trypanosoma vivax: a simplified protocol for in vivo growth, isolation and cryopreservation. Parasite. 2004;11:103–106. doi: 10.1051/parasite/2004111103. [DOI] [PubMed] [Google Scholar]
- Ohashi-Suzuki M., Yabu Y., Ohshima S., Nakamura K., Kido Y., Sakamoto K., Kita K., Ohta N., Suzuki T. Differential kinetic activities of glycerol kinase among African trypanosome species: phylogenetic and therapeutic implications. J. Vet. Med. Sci. 2011;73:615–621. doi: 10.1292/jvms.10-0481. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., de Godoy L.M.F., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteom. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Osório A.L.A.R., Madruga C.R., Desquesnes M., Soares C.O., Ribeiro L.R.R., Da Costa S.C.G. Trypanosoma (Duttonella) vivax: its biology, epidemiology, pathogenesis, and introduction in the new world – a review. Mem. Inst. Oswaldo Cruz. 2008;103:1–13. doi: 10.1590/S0074-02762008000100001. [DOI] [PubMed] [Google Scholar]
- Pereira C.A., Alonso G.D., Torres H.N., Flawiá M.M. Arginine kinase: a common feature for management of energy reserves in African and American flagellated trypanosomatids. J. Eukaryot. Microbiol. 2002;49:82–85. doi: 10.1111/j.1550-7408.2002.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yılmaz Ş., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A.F., Ternent T., Brazma A., Vizcaíno J.A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone T.M., Gonzatti M.I., Villamizar G., Escalante A., Aso P.M. Molecular profiles of Venezuelan isolates of Trypanosoma sp. by random amplified polymorphic DNA method. Vet. Parasitol. 2009;161:194–200. doi: 10.1016/J.VETPAR.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Perrone T., Aso P., Mijares A., Holzmuller P., Gonzatti M., Parra N. Comparison of infectivity and virulence of clones of Trypanosoma evansi and Trypanosoma equiperdum Venezuelan strains in mice. Vet. Parasitol. 2018;253:60–64. doi: 10.1016/j.vetpar.2018.02.024. [DOI] [PubMed] [Google Scholar]
- Pinto Torres J.E., Goossens J., Ding J., Li Z., Lu S., Vertommen D., Naniima P., Chen R., Muyldermans S., Sterckx Y.G.-J., Magez S. Development of a nanobody-based lateral flow assay to detect active Trypanosoma congolense infections. Sci. Rep. 2018;8:9019. doi: 10.1038/s41598-018-26732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirofski L., Casadevall A. Q&A: What is a pathogen? A question that begs the point. BMC Biol. 2012;10:6. doi: 10.1186/1741-7007-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe D.A., Belosevic M. Antibodies that recognize α- and β-tubulin inhibit in vitro growth of the fish parasite Trypanosoma danilewskyi, Laveran and Mesnil, 1904. Dev. Comp. Immunol. 2006;30:685–697. doi: 10.1016/J.DCI.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Portman N., Gull K. The paraflagellar rod of kinetoplastid parasites: from structure to components and function. Int. J. Parasitol. 2010;40:135–148. doi: 10.1016/j.ijpara.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanska M., Magez S., Dumont N., Pays A., Nolan D., Pays E. Antibodies raised against the flagellar pocket fraction of Trypanosoma brucei preferentially recognize HSP60 in cDNA expression library. Parasite Immunol. 2000;22:639–650. doi: 10.1046/j.1365-3024.2000.00348.x. [DOI] [PubMed] [Google Scholar]
- Radwanska M., Magez S., Michel A., Stijlemans B., Geuskens M., Pays E. Comparative analysis of antibody responses against HSP60, invariant surface glycoprotein 70, and variant surface glycoprotein reveals a complex antigen-specific pattern of immunoglobulin isotype switching during infection by Trypanosoma brucei. Infect. Immun. 2000;68:848–860. doi: 10.1128/IAI.68.2.848-860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston K.S., Hill K.L. The flagellum of Trypanosoma brucei: New tricks from an old dog. Int. J. Parasitol. 2008;38:869–884. doi: 10.1016/j.ijpara.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Barrios R.A., Valera Z., Parra O., Chacin E., Tavares-Marques L., Holzmüller P., Martínez-Moreno Á., Reyna-Bello A. Immunoreactive proteins of Trypanosoma vivax. Rev. Cientif. FCV-LUZ. 2015;25:311–316. [Google Scholar]
- Ramírez-Iglesias J.R., Eleizalde M.C., Reyna-Bello A., Mendoza M. Molecular diagnosis of cattle trypanosomes in Venezuela: evidences of Trypanosoma evansi and Trypanosoma vivax infections. J. Parasit. Dis. 2017;41:450–458. doi: 10.1007/s12639-016-0826-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez E., Perrone T., Recchimuzzi G., Cardozo I., Biteau N., Aso P., Mijares A., Baltz T., Berthier D., Balzano-Nogueira L., Gonzatti M. Molecular characterization and classification of Trypanosomaspp. Venezuelan isolates based on microsatellite markers and kinetoplast maxicircle genes. Parasit. Vectors. 2015;8:536. doi: 10.1186/s13071-015-1129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravia N.G., Hazbón M.H., Osorio Y., Valderrama L., Walker J., Santrich C., Cortázar T., LeBowitz J.H., Travi B.L. Protective immunogenicity of the paraflagellar rod protein 2 of Leishmania mexicana. Vaccine. 2005;23:984–995. doi: 10.1016/J.VACCINE.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Sasse R., Gull K. Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell. Sci. 1988;90:577–589. doi: 10.1242/jcs.90.4.577. [DOI] [PubMed] [Google Scholar]
- Shin Y., Lee Eung-goo, Shin G., Kim Young-rim, Lee Eun-young, Kim J., Jang H., Gershwin L.J., Kim D., Kim Yong-hwan, Kim G., Suh M., Jung T. Identification of antigenic proteins from Neospora caninum recognized by bovine immunoglobulins M, E, A and G using immunoproteomics. Proteomics. 2004;4:3600–3609. doi: 10.1002/pmic.200400963. [DOI] [PubMed] [Google Scholar]
- Siqueira-Neto J.L., Debnath A., McCall L.-I., Bernatchez J.A., Ndao M., Reed S.L., Rosenthal P.J. Cysteine proteases in protozoan parasites. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stijlemans B., Radwanska M., De Trez C., Magez S. African trypanosomes undermine humoral responses and vaccine development: link with inflammatory responses? Front. Immunol. 2017;8:582. doi: 10.3389/fimmu.2017.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez C., García F., Román D., Coronado A., Perrone T., Reyna A., Parra N. Factores de riesgo asociados a la tripanosomosis bovina en explotaciones ganaderas de Venezuela. Zootec. Trop. 2009;27:363–372. [Google Scholar]
- Taylor K.A. Immune responses of cattle to African trypanosomes: protective or pathogenic? Int. J. Parasitol. 1998;28:219–240. doi: 10.1016/S0020-7519(97)00154-9. [DOI] [PubMed] [Google Scholar]
- Taylor K.A., Mertens B. Immune response of cattle infected with African trypanosomes. Mem. Inst. Oswaldo Cruz. 1999;94:239–244. doi: 10.1590/S0074-02761999000200022. [DOI] [PubMed] [Google Scholar]
- Toro M., Leon E., Ruiz A. Resultados de un muestreo sobre tripanosomiasis bovina mediante técnicas serológicas. Vet. Trop. 1980;5:43–50. [Google Scholar]
- Valera Z., Parra O., Alvarado M., Barboza G., Escalona F., Ramirez R. Effect of experimental Trypanosoma vivax infection on hematological parameters in sheep. Rev. Cientif. FCV-LUZ. 2005;15:412–420. [Google Scholar]
- Verdillo J.C.M., Lazaro J.V., Abes N.S., Mingala C.N. Comparative virulence of three Trypanosoma evansi isolates from water buffaloes in the Philippines. Exp. Parasitol. 2012;130:130–134. doi: 10.1016/j.exppara.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Werbovetz K.A. Tubulin as an antiprotozoal drug target. Mini Rev. Med. Chem. 2002;2:519–529. doi: 10.2174/1389557023405648. [DOI] [PubMed] [Google Scholar]
- Werbovetz K.A., Sackett D.L., Delfín D., Bhattacharya G., Salem M., Obrzut T., Rattendi D., Bacchi C. Selective antimicrotubule activity of N1-phenyl-3,5-dinitro-N4,N4-di-n-propylsulfanilamide (GB-II-5) against kinetoplastid parasites. Mol. Pharmacol. 2003;64:1325–1333. doi: 10.1124/mol.64.6.1325. [DOI] [PubMed] [Google Scholar]
- Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wilson K., Berens R.L., Sifri C.D., Ullman B. Amplification of the inosinate dehydrogenase gene in Trypanosoma brucei gambiense due to an increase in chromosome copy number. J. Biol. Chem. 1994;269:28979–28987. [PubMed] [Google Scholar]
- Woo P.T. The haematocrit centrifuge for the detection of trypanosomes in blood. Can. J. Zool. 1969;47:921–923. doi: 10.1139/z69-150. [DOI] [PubMed] [Google Scholar]
- Worku N., Stich A., Daugschies A., Wenzel I., Kurz R., Thieme R., Kurz S., Birkenmeier G. Ethyl pyruvate emerges as a safe and fast acting agent against Trypanosoma brucei by targeting pyruvate kinase activity. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S.C., Kumar Ritesh, Kumar J., Singh M., Bera B.C., Kumar Rajender, Tatu U., Tehri K. Antigenic characterization of 52–55 kDa protein isolated from Trypanosoma evansiand its application in detection of equine trypanosomosis. Res. Vet. Sci. 2017;114:455–460. doi: 10.1016/j.rvsc.2017.07..1016/j.rvsc.2017.07.034. [DOI] [PubMed] [Google Scholar]
- Yakovich A.J., Ragone F.L., Alfonzo J.D., Sackett D.L., Werbovetz K.A. Leishmania tarentolae: purification and characterization of tubulin and its suitability for antileishmanial drug screening. Exp. Parasitol. 2006;114:289–296. doi: 10.1016/j.exppara.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.