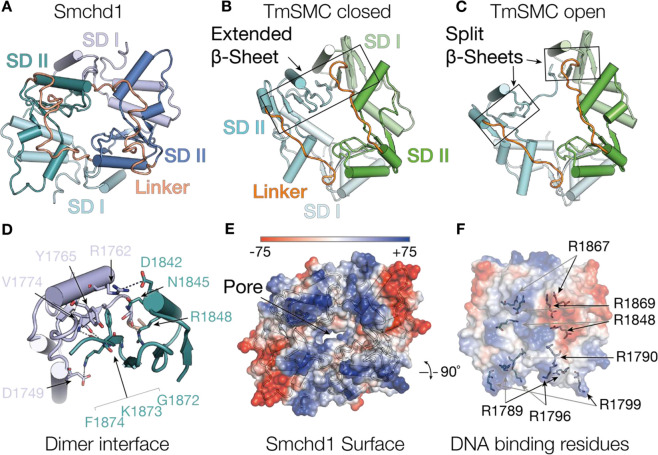
Figure 3. The Smchd1 Hinge domain.
(A) Structure of the mouse Smchd1 hinge domain showing cartoon representation of the two protomers (green and blue) in the homodimer indicating subdomain I (SD I, pale colours) and subdomain II (SD II, dark colours) and subdomain linker region (orange). (B,C) Structures of the Thermotoga maritima SMC hinge domain showing both protomers (cyan and green) in the dimers, subdomain I (SD I, pale colours) and subdomain II (SD II, dark colours) and subdomain linker region (orange) displayed. (B) shows a closed hinge domain conformation with an extended β-sheet and (C) open conformation with a split β-sheet at one dimerisation interface. Structures are displayed in the same orientation as Smchd1, aligning on subdomain II of the blue and green protomers from smchd1 and T. maritima SMC hinge domains, respectively. (D) Cartoon representation of the Smchd1 HD dimerisation interface between SD I (pale) of one protomer to SDII from the opposing protomer (colouring as in panel (A)). Key residues involved in the interaction are indicated in stick representation with hydrogen bonding and electrostatic interactions indicated with dashed lines. (E) Surface electrostatic representation of the Smchd1 HD indicating the surface exposed positive region in the interdomain linker region. (F) Surface representation of the Smchd1 HD showing positively charged arginine residues involved in binding DNA. These include surface exposed residues from the interdomain linker region (R1789, R1796, R1799) and residues buried in the dimer pore (R1790, R1848, R1869), and R1867, which is substituted to glycine in an FSHD patient kindred. Black and grey arrows indicate residues from opposing protomers in the Smchd1 dimer.