Fig. 3. RCAN1 TxxP and SPP motifs inhibit CN by binding and blocking the active site.
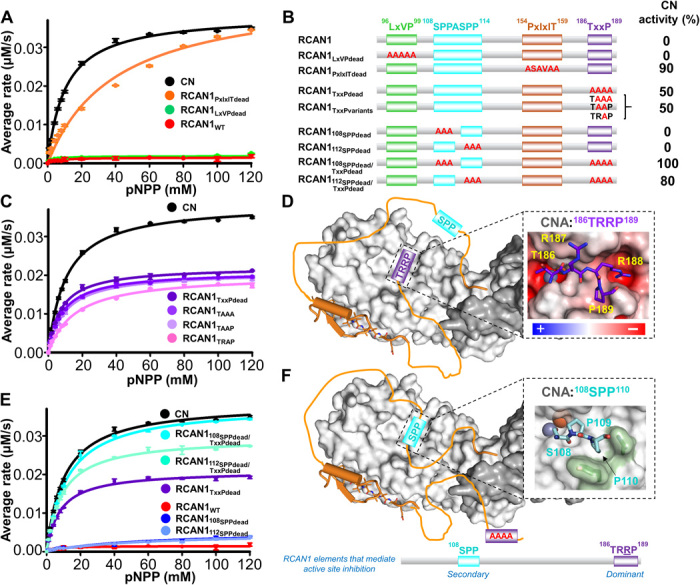
(A) Enzymatic activity of CN in the absence (black) and presence of RCAN1 (red) and its SLiM-binding motif variants (PxIxITdead, orange; LxVPdead, green); pNPP assays; ±SE, n = 3. (B) RCAN1 domain diagram illustrating the RCAN1 mutants tested with the corresponding CN activity (relative to free CN). (C) Same as (A) but with RCAN1 TxxP variants. (D) Cartoon illustrating how the TxxP motif engages the CN active site. Inset: Model of the RCAN1 TxxP (TRRP; purple) motif bound to CN based on the structure of the CNA AID domain (27); CN is shown as an electrostatic potential energy surface. (E) Same as (A) but with RCAN1 SSPASSP and TxxP variants. (F) Same as (D), illustrating CN inhibition in the absence of a functional TxxP motif. Inset: Model of the RCAN1 108SPP (cyan) motif bound to CN based on the structure of the CNA AID domain.
