Fig. 6. RCAN1 is both a potent inhibitor of CN and a substrate.
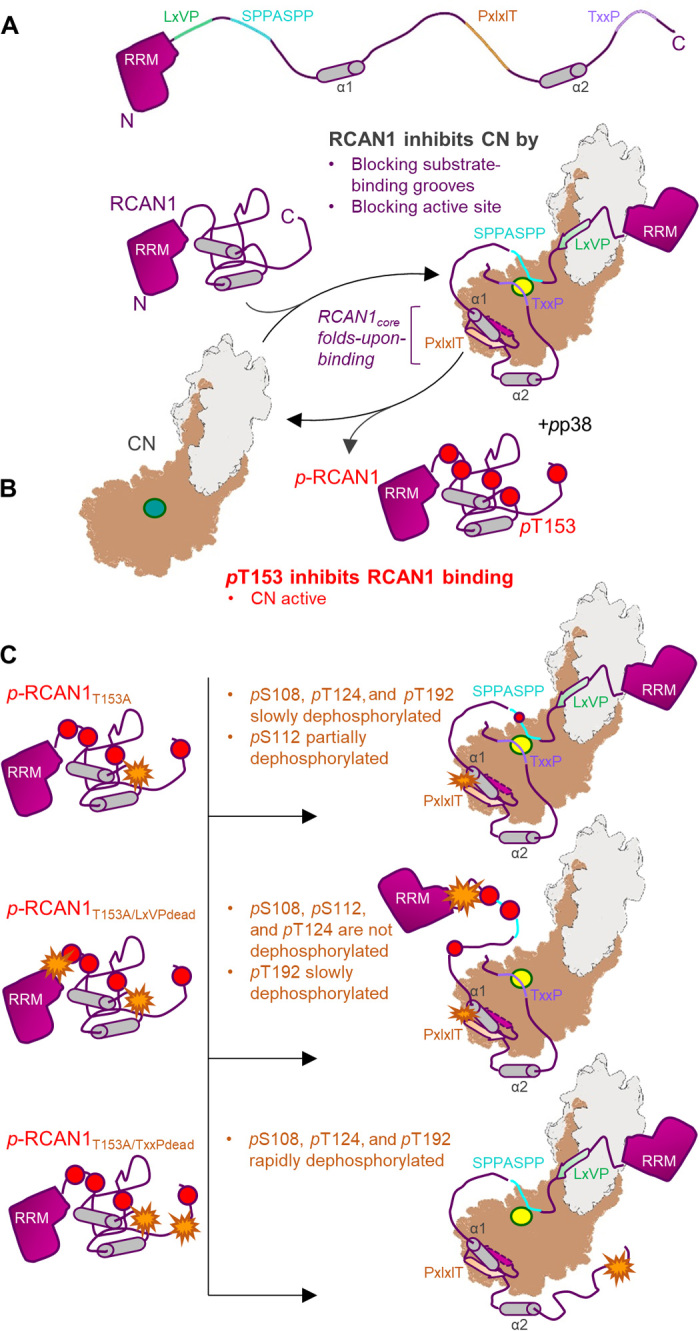
(A) RCAN1 is a potent CN inhibitor. RCAN1 (magenta, with key sequence and structural features indicated) binds and inhibits CN (gray/beige; active site in yellow) via two mechanisms. First, RCAN1 blocks CN-specific substrate LxVP and PxIxIT interaction grooves by binding these pockets using its LxVP and especially its PxIxIT motifs. This prevents canonical CN substrates, like the NFATs, from binding CN. Second, RCAN1 directly blocks the CN active site via its TxxP motif and, to a lesser extent, its SPPASSP motif. This further reduces CN activity against its endogenous substrates. The RCAN1core folds upon binding CN. (B) Phosphorylation of RCAN1 T153 (pT153) inhibits CN binding. Active p38 phosphorylates RCAN1 on pS108, pS112, pT124, pT153, and pT192. Phosphorylation of RCAN1 T153 (pT153), which is immediately N-terminal to the PxIxIT motif, lowers the affinity of RCAN1 for CN, leading to dissociation of the complex and increased CN activity. (C) Phosphorylated RCAN1 (pRCAN1) is weakly dephosphorylated by CN. In the absence of phosphorylation at T153 (pRCAN1T153A; T153A represented as an orange start), pRCAN1 is able to bind CN with strong affinity. This results in the slow dephosphorylation of pS108, pT124, and pT192, with pS112 becoming partially dephosphorylated (top). However, with the exception of pT192, these dephosphorylation events require the presence of the nearby LxVP motif. That is because, in the LxVPdead mutant (T153A and LxVPdead represented as orange stars), pS108, pS112, and pT124 remain phosphorylated, even after 20 hours (middle). Last, if the inhibitory TxxP sequence is inactivated (TxxPdead; T153A and TxxPdead represented as orange stars), the ability of CN to dephosphorylate pRCAN1 is substantially enhanced, with dephosphorylation rates increasing ~10-fold (bottom).
