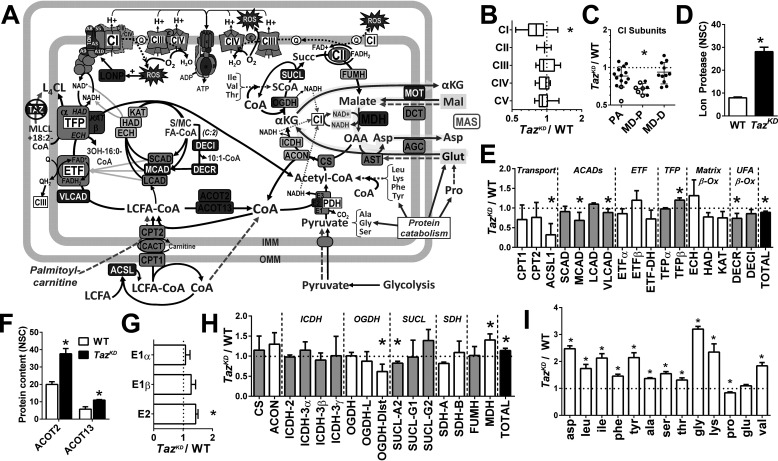
Figure 5.
Remodeling of the mitochondrial proteome in the Taz-deficient heart. A, summary schematic illustrating enzymes (boxes) found by LC–MS proteomic profiling of cardiac mitochondria to be higher (red fill), lower (blue fill), or not different (gray fill) in TazKD compared with WT mice (q < 0.05; n = 3/group). Metabolites detected by GC/MS in snap-frozen cardiac tissue are shown in red (TazKD > WT), blue (TazKD < WT), black (TazKD = WT), or gray font (not measured, but included for context) (q < 0.05; n = 4–6/group). Green dashed arrows indicate entrance sites of exogenous substrates added in mitochondrial respiration experiments, with glutamate oxidation through the MAS highlighted in yellow. B–I, graphical data representing the relative (TazKD/WT) expression of cumulative normalized spectral counts corresponding to: respiratory chain complexes (B); complex 1 subunits localized to the peripheral arm (PA), proximal (MD-P), and distal membrane domain (MD-D) (C); Lon protease (LONP) (D); fatty acid oxidation enzymes (E); acyl-CoA thioesterases (ACOT) 2 and 13 (F); pyruvate dehydrogenase (PDH) complex subunits (G); tricarboxylic acid cycle enzymes (H); and cardiac free amino acid content (per mg tissue protein) detected by GC/MS (I). *, q < 0.05 TazKD versus WT. ACADs, acyl-CoA dehydrogenases; ACON, aconitase; ACSL, acyl-CoA synthetase; AGC, aspartate-glutamate cotransporter; AST, aspartate transaminase; CACT, carnitine-acyl-CoA translocase; CS, citrate synthase; DCT, dicarboxylate transporter; DECR, dienoyl-CoA reductase; DECI, dienoyl-CoA isomerase; ECH, enoyl-CoA hydratase; ETF, electron transferring flavoprotein; FUMH, fumarate hydratase; HAD, hydroxyacyl-CoA dehydrogenase; ICDH, isocitrate dehydrogenase; KAT, ketoacyl-CoA thiolase; OGDH, oxoglutarate dehydrogenase; LCFA, long-chain fatty acid; MOT, malate-oxogluarate transporter; SUCL, succinyl-CoA ligase.