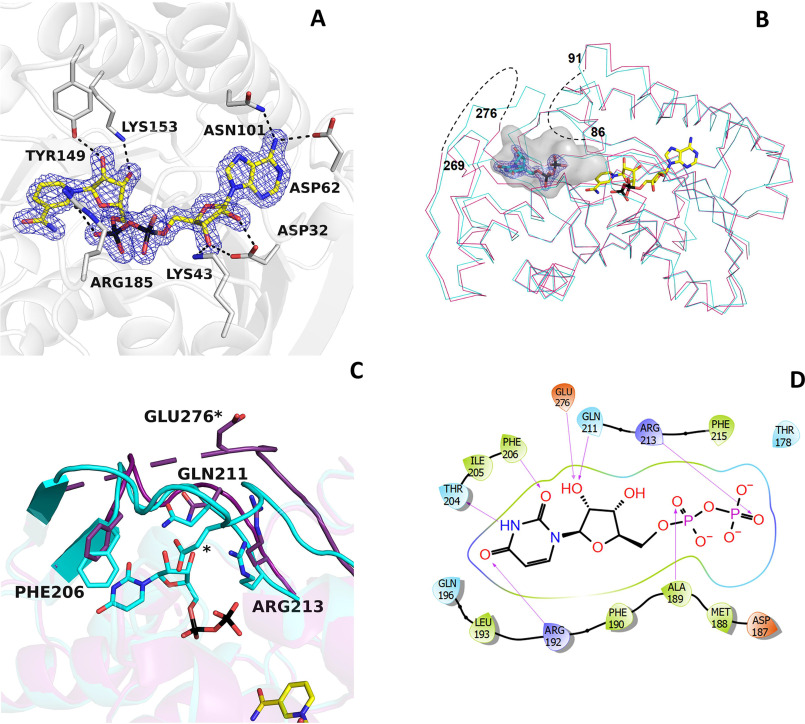
Figure 2.
NAD+ and UDP binding in BcUGAepi. A, weighted 2Fo − Fc electron density of NAD+ in the epimerase/NAD+ binary complex (subunit A). The contour level is 1.4σ. Side chains involved in hydrogen-bonding and electrostatic interactions are shown (light gray carbons). Together with Thr-126 (Fig. 3), Tyr-149, and Lys-153 are part of the fingerprint catalytic triad of the SDR family. B, superposition between the Cα traces of the epimerase structures bound to NAD+ (purple) and NAD+/UDP (cyan). NAD+ carbons are shown in yellow, and UDP carbons are shown in cyan. The electron density of UDP is shown with a contour level of 1.4σ. The loops 86–91 and 269–276 are disordered in the NAD+ complex (dashed lines). Upon UDP binding, both loops adopt an ordered conformation that defines a large cavity where the mononucleotide ligand binds. The large cavity space between the UDP pyrophosphate and the nicotinamide forms the sugar-binding site. C, ribbon diagram of the NAD+ structure (purple) superposed onto the complex with NAD+/UDP (cyan). In the absence of UDP, loop 269–276 is disordered. UDP binding reorganizes this loop that, together with residues 204–214, wraps the UDP molecule. D, two-dimensional schematic diagram of the extensive interactions between UDP and the protein residues.