Figure 3.
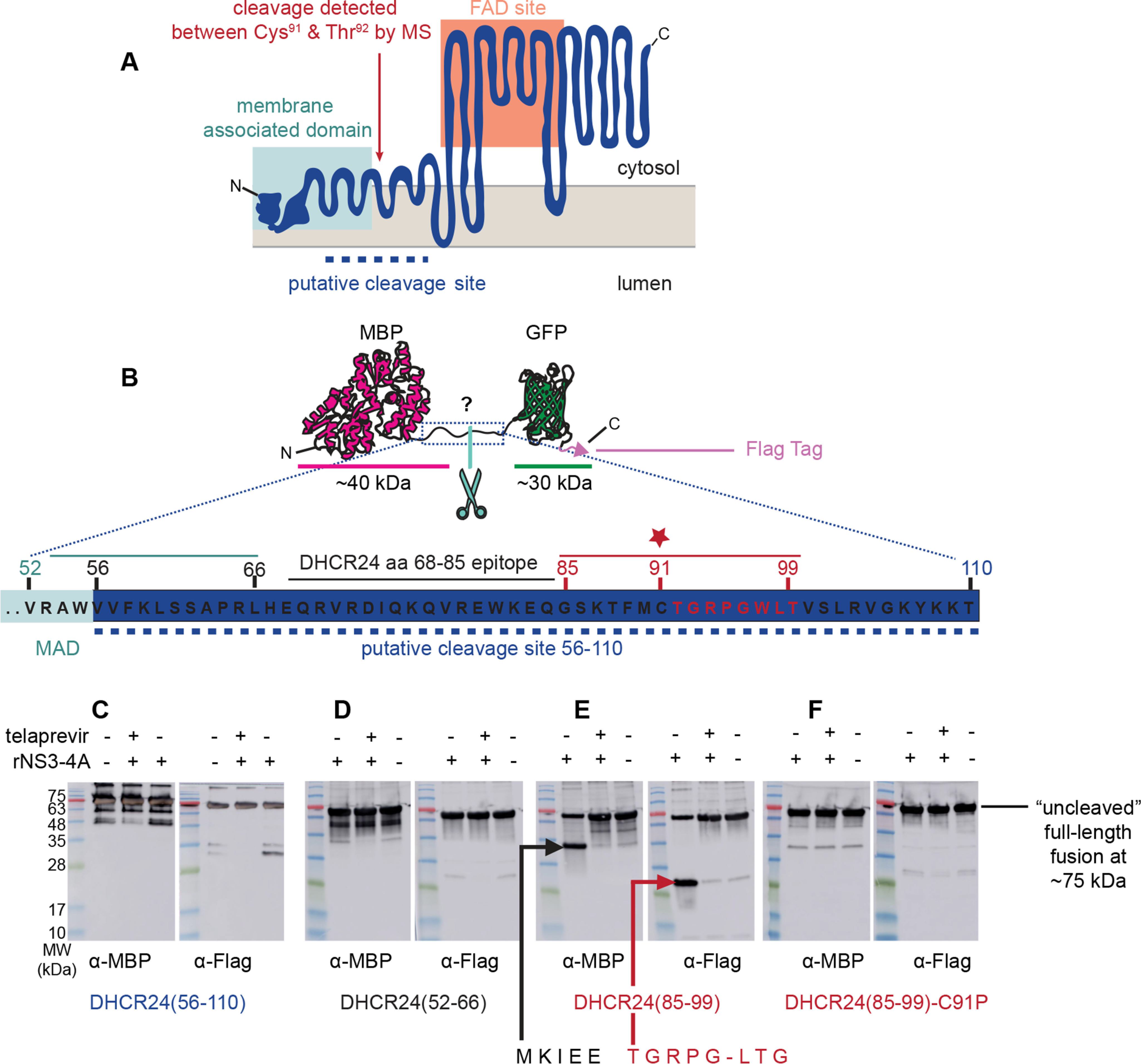
DHCR24 is proteolytically cleaved by NS3-4A between Cys91 and Thr92. A, the topology model of DHCR24 predicts (54) an N-terminal membrane-associated domain that tethers a C-terminal FAD-binding site to the surface of the ER membrane. The dashed line indicates the region suggested by antibody reactivity to contain the cleavage site. Antibody mapping of the cleavage products from in vitro reaction of recombinant DHCR24–FLAG and NS3-4A proteins predicted cleavage between the N- and C-terminal domains. The experimentally mapped cleavage site between Cys91 (C91) and Thr92 (T92) is indicated. B, to map the cleavage site, peptides spanning different regions of the putative cleavage region were cloned between an N-terminal MBP domain and a C-terminal GFP domain with a FLAG tag. C, incubation of the candidate substrate containing DHCR24 residues 56–110 with recombinant NS3-4A produced a faster-migrating band when probed with an anti-FLAG antibody. This was blocked in the presence of telaprevir, an NS3-4A inhibitor. Analogous reactions were performed with test substrates containing DHCR24 residues 52–66 or 85–99. D, no cleavage was observed for the DHCR24(52–66) substrate. E, cleavage of the DHCR24(85–99) substrate by recombinant NS3-4A was detectable by immunoblot using both anti-MBP and anti-GFP. Edman degradation of the GFP-containing peptide revealed the amino acid sequence of NH2-TGRPGLTG-COOH, indicating that cleavage occurs between Cys91 and Thr92. F, cleavage of the DHCR24(85–99) substrate was abrogated by mutation of Cys91 to Pro. Note that the peptide spanning map in B and the immunoblot in E are reproduced in Fig. S5 to help orient the reader in viewing the Edman degradation traces. MW, molecular mass.
