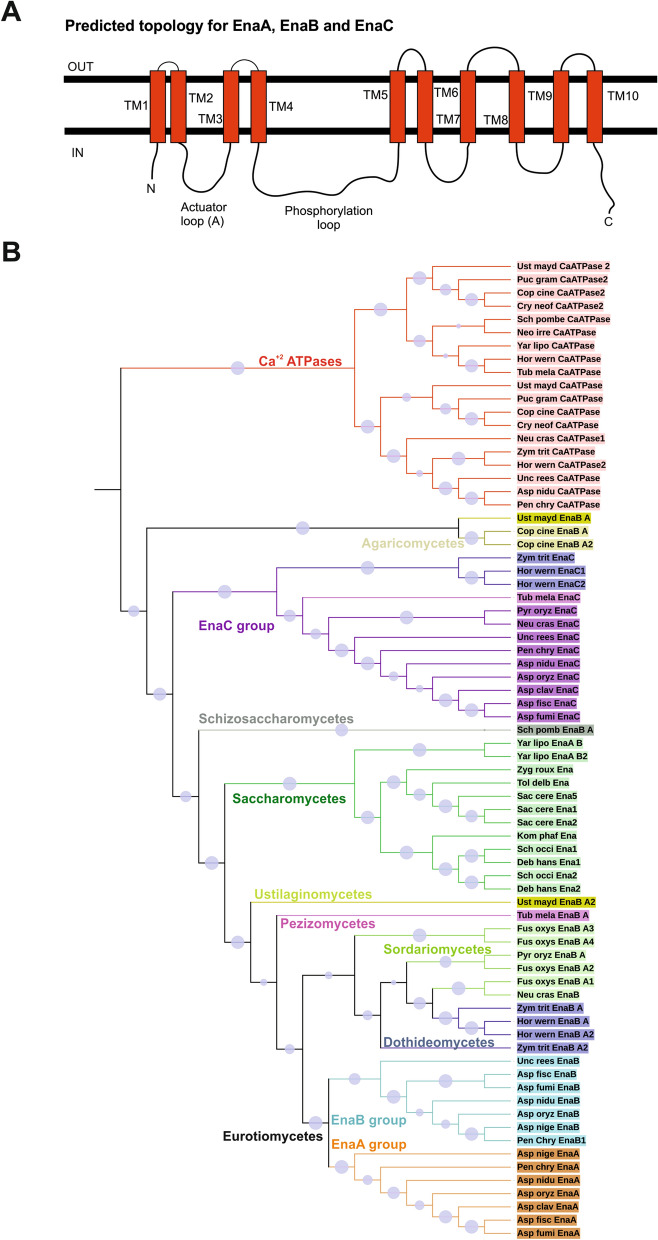
Figure 1.
ENA-like proteins in Aspergillus nidulans. (A) Proposed two-dimensional model of EnaA and EnaB describing the predicted membrane topology and principal conserved functional domains among P-type ATPases. (B) Maximum-likelihood tree of Na+, K+, H+ ATPases from fungi and yeast. EnaA and EnaB clustered in the same group of ATPases while EnaC clearly differentiated and belong to a separate clade. MEGA7 software was used to generate the tree58 and it was edited with iTOL59. Blue circles in each clade represent bootstrap values. Species; Asp_clav: Aspergillus clavatus; Asp_fisc: Aspergillus fischeri; Asp_fumi: Aspergillus fumigatus; Asp_nidu: Aspergillus nidulans; Asp_nige: Aspergillus niger; Asp_oryz: Aspergillus oryzae; Deb_hans: Debariomyces hansenii; Fus_oxys: Fusarium oxysporum; Hor_wern: Hortaea werneckii; Kom_pahf: Komagataella phafii; Neu_cras: Neurospora crassa; Pen_chry_ Penicillium chrysogenum; Pyr_oryz_ Pyricularia oryzae; Sac_cere: Saccharomyces cerevisiae; Sch_occi: Schwanniomyces occidentalis; Tol_delb: Torulaspora delbrueckii; Unc_rees: Uncinocarpus reesii; Ust_mayd: Ustilago Maydis; Zyg_roux: Zygosaccharomyces rouxii. Cop_cine: Coprinopsis cinerea; Cry_neof: Cryptococcus neoformans; Puc_gram: Puccinia graminis; Tub_mela: Tuber melanosporum; Yar_lipo: Yarrowia lipolytica; Sch_pomb: Schizosaccharomyces pombe; Neo_irre: Neolecta irregularis; Zym_trit: Zymoseptoria tritici.