Fig. 3.
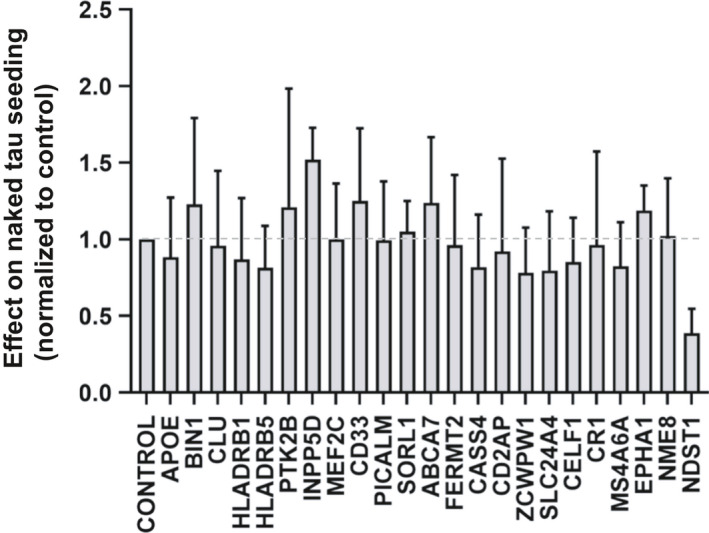
Knockout of AD GWAS genes does not modify naked tau seeding. GWAS genes were individually targeted in HEK293T RD(P301S)‐C/CL biosensor cells using CRISPR/Cas9 to create polyclonal knockout cell lines, which were cultured for 2 weeks in the presence of puromycin. Recombinant tau fibrils were added to those cells to induce seeding. Seeding was quantified using FRET, and the percentage of FRET‐positive cells was normalized to the scrambled gRNA. Data were collected in triplicate. The X‐axis indicates the targeted genes, and the Y‐axis indicates normalized seeding activity. None of the genes modified the seeding efficiency. Heparin and NDST1 were positive controls for uptake inhibition. Error bars indicate the SEM. The dotted gray line represents the value of control cells. One‐way ANOVA was used to analyze the result and test for statistically significant differences.
