Figure 5. The canonical high‐affinity ZEB1 motif is dispensable for the formation of a ZEB1 activator complex.
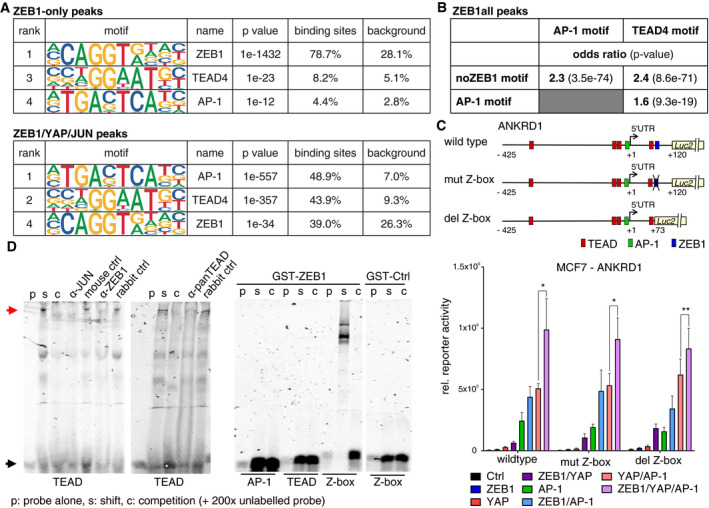
- Enrichment of the ZEB1, AP‐1 and TEAD4 DNA binding motifs is shown, as identified by HOMER known motif analysis on 200 bp regions centred on different subsets of ZEB1 peak summits. An extended list of the identified motifs can be found in Appendix Table S3 and S4.
- Hypergeometric testing showed that while there is a significant association between the AP‐1 and TEAD4 motif within ZEB1 peaks, both motifs are associated with the absence of the ZEB1 consensus motif (Z‐box). Odds ratio (OR): measure of association between categorical variables with OR > 1 indicating a positive association. P‐value from Fisher's exact test.
- The upper panel shows a schematic representation of the ANKRD1 luciferase reporter constructs. The lower panel shows luciferase reporter assays with these different ANKRD1 promoter constructs in MCF7 cells upon overexpression of ZEB1, YAP and JUN/FOSL1 or different combinations of the factors. n = 4–5; shown is mean ± s.e.m.; *P ≤ 0.05; **P ≤ 0.01; ratio paired t‐test. Firefly luciferase activity was normalised to co‐transfected Renilla luciferase. The wild‐type assay corresponds to the assay shown in Fig 4E.
- Left panel: EMSA using nuclear extracts from MDA‐MB‐231 and labelled TEAD binding side as probe (black arrow). Competition of specific binding complex (read arrow) with cold probe (C), anti‐JUN, ‐panTEAD, and ‐ZEB1 antisera. Right panel: recombinant GST‐ZEB1, but not GST‐Ctrl, only binds to Z‐box.
