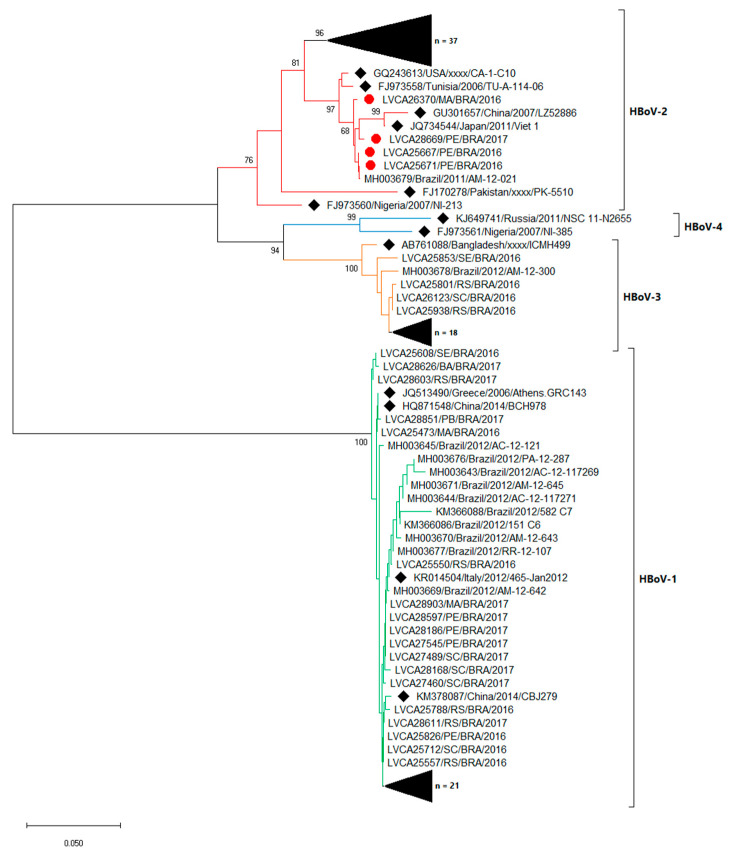
Figure 4.
Phylogenetic analyses based on VP1 nucleotide (nt) sequences of circulating Brazilian human bocavirus (HBoV) strains. Reference strains, marked with a black filled diamond, were downloaded from GenBank and labelled with their accession number followed by country, year and register number. Strains obtained are shown as per LVCA followed by the internal register number, state, country and year of collection (i.e., LVCA28597/PE/BRA/2017), and the red filled circle represents sequences with a three-nt insertion. Neighbour-joining phylogenetic tree was constructed with MEGA X software and bootstrap tests (2000 replicates), based on the Kimura two-parameter model. Bootstrap values above 70% are given at branch nodes.