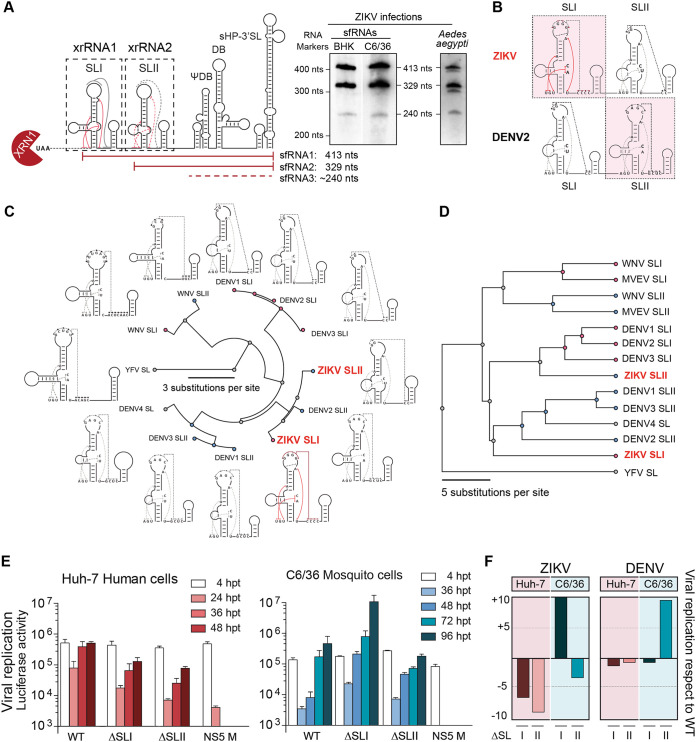
FIG 3.
Distinct functions of duplicated stem-loop structures present at the ZIKV 3′ UTR. (A) Schematic representation of ZIKV 3′ UTR showing the two XRN1-resistant RNAs involved in sfRNA formation (xrRNA1 and xrRNA2, including SLI and SLII, respectively). At the right are Northern blots showing the pattern of sfRNAs generated during ZIKV infection in vertebrate and invertebrate cells and A. aegypti mosquitoes as indicated. The two main sfRNAs of 413 and 329 nucleotides are shown together with a minor sfRNA of about 240 nucleotides. (B) Comparison of secondary structures of duplicated SLs from ZIKV and DENV2 based on available biochemical probing and crystal structures. (C) Fan dendrogram indicating the distance between SL structures of the indicated mosquito-borne flaviviruses (MBFVs). SLI and SLII of different MBFVs correspond to the structures included in xrRNA1 and xrRNA2, respectively, and their locations (first and second) are with respect to the 5′ end of the 3′ UTR. (D) Phylogenetic analysis based on sequence alignments of SLs from different MBFVs. (E) Distinct functions of the two SLs in human and mosquito cells, as indicated in each case. Replication of recombinant reporter ZIKV carrying deletions of xrRNA1 (ΔSLI) or xrRNA2 (ΔSLII) is shown together with the WT and a replication-impaired control NS5 M as a function of time. The luciferase values are means ± standard deviations of a representative experiment (three independent experiments were performed). (F) Roles of SLI and SLII of ZIKV and DENV in viral replication in the two hosts support inverted location of these two RNA elements.