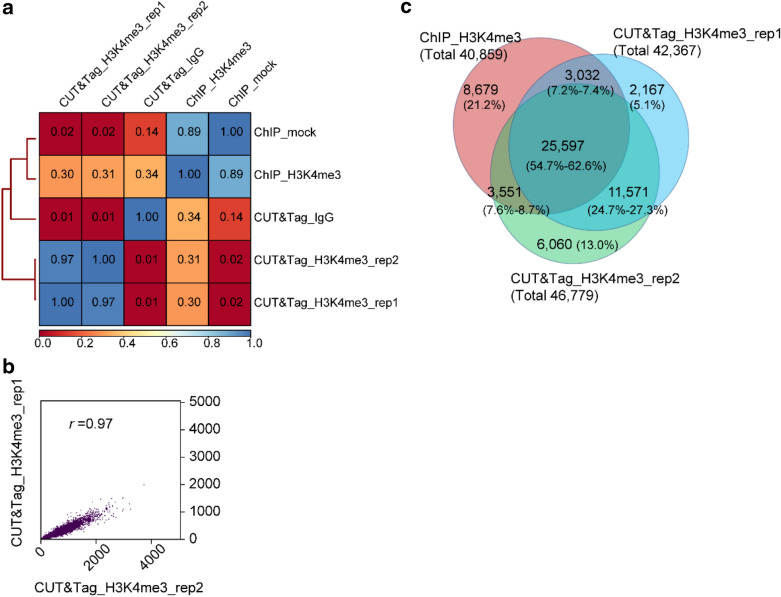
Fig. 3.
Correlation analysis of CUT&Tag and ChIP samples. a Hierarchically clustered correlation matrix of CUT&Tag replicates (rep1 and rep2) and with ChIP-seq profiling for the H3K4me3 histone modification. The same antibody was used in all experiments. Pearson correlations were calculated in deepTools (the multiBamSummary was followed with plotCorrelation tools) using the read counts split into 500-bp bins across the genome. b Scatterplot correlation of CUT&Tag replicates (rep1 and rep2). Pearson’s r was indicated. (c) Number of shared peaks and unique peaks in CUT&Tag replicates (rep1 and rep2) and ChIP-seq. Peaks were called by macs2 using randomly sampled 6-M clean data of CUT&Tag and 24-M clean data of ChIP. Peaks overlapped across the genome and with the distance of peak summit < 300 bp were considered as the same peak