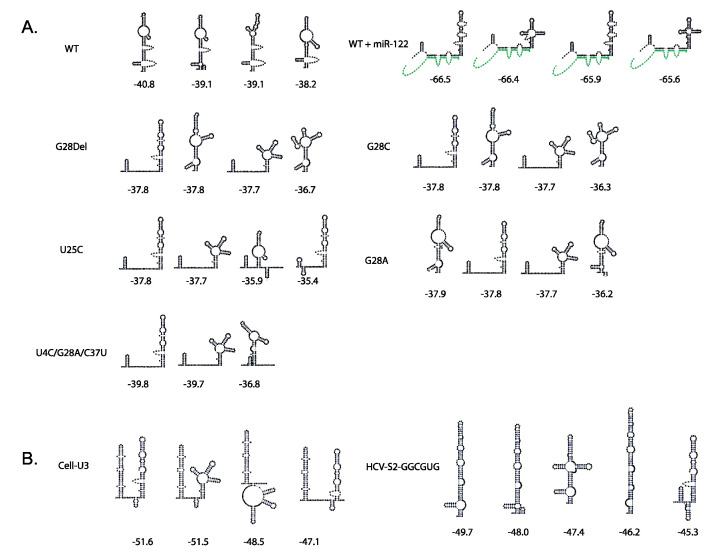
Figure 3.
Secondary structure predictions of HCV mutants showing formation of SLII. (A) In silico secondary structure predictions (experimentally unvalidated structures) for the domain I and domain II of wild type HCV with and without miR-122 [21] and mutant RNA sequences of viruses capable of miR-122-independent replication [23,45,46]. One or more RNA structures show formation of translationally active IRES SLII structure for each mutant. (B) We performed secondary structure predictions using an online software ‘RNAstructure’ of Cell-U3 [50] and HCV-S2 GGCGUG [66] RNA sequences. These predicted structures showed formation of SLII in one or more structures which may be due to formation of another short stem loop upstream of SLII.