Abstract
Downy mildew, powdery mildew, and grey mold are some of the phytopathological diseases causing economic losses in agricultural crops, including grapevine, worldwide. In the current scenario of increasing global warming, in which the massive use of agrochemicals should be limited, the management of fungal disease has become a challenge. The knowledge acquired on candidate resistant (R) genes having an active role in plant defense mechanisms has allowed numerous breeding programs to integrate these traits into selected cultivars, even though with some limits in the conservation of the proper qualitative characteristics of the original clones. Given their gene-specific mode of action, biotechnological techniques come to the aid of breeders, allowing them to generate simple and fast modifications in the host, without introducing other undesired genes. The availability of efficient gene transfer procedures in grapevine genotypes provide valid tools that support the application of new breeding techniques (NBTs). The expertise built up over the years has allowed the optimization of these techniques to overexpress genes that directly or indirectly limit fungal and oomycetes pathogens growth or silence plant susceptibility genes. Furthermore, the downregulation of pathogen genes which act as virulence effectors by exploiting the RNA interference mechanism, represents another biotechnological tool that increases plant defense. In this review, we summarize the most recent biotechnological strategies optimized and applied on Vitis species, aimed at reducing their susceptibility to the most harmful fungal and oomycetes diseases. The best strategy for combating pathogenic organisms is to exploit a holistic approach that fully integrates all these available tools.
Keywords: plant biotechnology, Vitis vinifera, RNA interference, genome editing, biotic stresses
1. Introduction
Grapevine is one of the world’s most commonly produced fruit crops, with a yield of about 79 million tons of grapes produced only in 2018 (Faostat Database, 2018). High-quality grapes for table consumption and wine production are derived from varieties of only one vine species, Vitis vinifera L. subsp. sativa, whereas other species are exploited as rootstocks [1], or are used in breeding programs solely for introducing new important traits in selected cultivars [2]. However, this kind of application is somewhat controversial due to international rules, particularly in Europe, where a limitation is imposed on the use of cultivars derived only from Vitis vinifera within breeding programs [3].
Considering their high pedoclimatic adaptation capacity, the cultivation of Vitis vinifera cultivars become possible between 30° to 50 °N and S latitude [4]. In the presence of favorable weather conditions (generally mild temperatures and high humidity), during the crop cycle, almost every organ of the plant is susceptible to attack by the main fungal and oomycetes diseases, such as downy mildew, powdery mildew, and grey mold, caused by Plasmopara viticola (Berk. and Curtis) Berl. and De Toni, Erysiphe necator Schwein., and Botrytis cinerea Pers., respectively [5]. According to recent global surveys, these diseases in regards to the main winegrowing regions in the world were considered by researchers and production professionals as the most harmful for grape production [6,7,8,9].
Grapevine breeding programs are mainly focused on inducing resistance against biotic agents, especially those that have a history of attacking European Vitis since the late nineteenth century, such as grape phylloxera and mildews. Mildews originated from North America, and they were introduced in most European Vitis vinifera varieties that proved to be highly susceptible due to the absence of coevolutionary processes between pathogens and plants [10]. Research work on resistant (R) genes and their introduction in selected cultivars for genetic improvement by classical breeding is generally challenging and requires several generations of backcrosses, during which a strict selection must be carried out, trying to synchronously preserve either important agronomic/oenological characteristics or traits of interest. The varietal rigidity imposed by registered designations of origin, long juvenile phase, and high heterozygosity leads to costly and longer breeding technical times when classical breeding is applied on Vitis species [11]. Furthermore, although classical breeding and agrochemical approaches were considered effective at first, in the long run, they could lead to the emergence of resistant pathogen strains, since they mainly confer a qualitative type of resistance which is prevalently monogenic [12]. To cope with these threats, farmers have massively used pesticides, arousing conflicting opinions regarding environmental sustainability and the quality of viticulture and wine production processes [13].
Studies in the field of plant molecular biology and biotechnology may support plant defense strategies, allowing researchers to select traits that could undermine the pathogen’s aggression [14]. Genetic transformation remains generally the most commonly exploited strategy compared to several other biotechnological approaches, as it allows researchers to stably insert specific gene sequences into a host plant. Genetic transformation also permits the importation of more than one R gene, and this creates the condition to have potential additive or synergistic effects. The validation of R genes in Vitis species is possible, but it requires the development of efficient regeneration and transformation protocols in order to genetically transform these plants, which often lead researchers to opt for model plants like Nicotiana tabacum or Arabidopsis thaliana, as hosts to implement these studies [15]. Classical genetic engineering techniques, mainly based on standard genetic transformation methods through the insertions and consequent expression of heterologous genes of interest, such as resistance (R) genes or other defense genes, represent the basis on which the new generation of biotechnologies are founded [11].
Overexpression of defense genes against crop fungal pathogens symbolizes one of the main biotechnological tools exploited to counterbalance pathogen aggressiveness, and consequent yield losses [14,16]. Pathogenesis-related proteins (PR proteins), antimicrobial peptides, secondary metabolites, and specific compounds can be overexpressed in host cells with a direct effect at the target level. Alternatively, it is also possible to stimulate host defense biosynthetic pathways (e.g., through the overexpression of transcription factors that enhance the plant defense-related genes) [17]. In addition to these biotechnological strategies, new breeding techniques (NBTs) such as genome editing mediated by CRISPR/Cas9 technology, a high precision tool capable of strategically introducing targeted mutations in the host genome [18], or cisgenesis/intragenesis which allow the inclusion of gene sequences from sexually compatible plants [19], have been developed and optimized during recent decades. The RNA interference (RNAi) mechanism, where double-strand RNA (dsRNA) molecules trigger the mRNA degradation or translational repression, is another powerful tool to subvert pathogenic attacks while the downregulation of gene expression occurs [20,21].
This literature review aims to provide an overview of target genes discovered in Vitis species and assessed through the abovementioned biotechnological strategies to increase tolerance to the most severe pathogens, such as grey mold, powdery and downy mildews.
2. Plant Response Mechanisms to Pathogenic Attacks
Plants have a series of biochemical or physical barriers that belong to the general constitutive defenses which can prevent fungi from entering the plant cells. Plant cell wall and related compounds such as trichomes, wax layers, cuticle, cellulose, and pectin lamellae are the main physical impediments to fungal ingression. For example, the presence of numerous trichomes on the lower leaf epidermis reduces downy mildew primary infection due to the increased exposure of zoospore to dehydration [22]. After infection, several histological responses help plant cells by curbing pathogen invasion. Callose deposition-forming cell wall thickenings, commonly known as papillae, lignin, and other phenolic compounds production nearby fungal penetration sites, have shown an active defense role during the early stages of plant invasion [23]. For instance, the expression of stress-induced callose synthase PMR4 (Powdery Mildew Resistance 4) has proven to provide complete penetration resistance against Arabidopsis powdery mildew [24].
During evolutionary-conserved plant defense processes, two effective and subsequent mechanisms occur to actively respond to pathogen and pest infections; firstly, the activation of the response machinery takes place following the perception of non-specific molecules produced by the pathogen; secondly, there occurs a specific recognition of pathogens virulence factors, through the products encoded by R genes [25]. Plants are able to generally recognize bacteria, fungi, oomycetes, and viruses thanks to either the presence or the production of specific conserved molecules, known as microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) that act as elicitors of plant defense responses [26]. In the case of pathogenic attack, the presence of some receptors and co-receptors known as pattern recognition receptors (PRRs) on the plant surface at the membrane level, can efficiently recognize PAMPs, thus, allowing the establishment of PAMP-triggered immunity (PTI) defense response, leading to the impediment of early-stage fungal growth, without killing the pathogen cells [27].
In order to suppress these barriers, pathogens can secrete a plethora of effectors which, in some cases, can be identified by the plant cell thanks to the presence of Resistance proteins (R proteins), which lead to the Effector-triggered Immunity (ETI) [28]. This plant immunity response is stronger than PTI, as it is able to elicit the activation of additional defense signaling mechanisms including PR genes expression induction, local hypersensitive responses (HR), and consequently, programmed cell death (PCD) [29]. Unfortunately, due to the lack of specific R genes, the most important Vitis vinifera cultivars have proved to have inadequate defense responses to limit the invasion of both biotrophic and necrotrophic Fungi and Chromista pathogens [5].
3. Genetic Engineering for the Expression of Candidate Genes Involved in Fungal-Oomycete Resistance
Prior to genetic transformation processes, a fundamental part is the identification of candidate genes that exert in the host an active role in the enhancement of plant defenses, such as pathogenesis-related proteins, antimicrobial peptides, transcriptional factors, products of the secondary metabolism, and defense-related genes. The expression/overexpression through genetic engineering techniques is still one of the most common biotechnological tools used to validate cisgenic and transgenic sequences that induce/improve resistance against specific pathogens in Vitis spp. (Table 1).
Table 1.
Representative attempts of genetic transformation applied in Vitis species to enhance resistance against the most harmful fungal and oomycetes pathogens.
| Gene Name | Gene Source | Description | Biological Effect | Host | References |
|---|---|---|---|---|---|
| Pathogenesis-Related Proteins | |||||
| Non-expressor of Pathogenesis Related 1 ( VvNPR1.1 ) | Vitis vinifera | Key signal in salicylic acid pathway and local basal resistance to biotrophs | Enhanced resistance to Erysiphe necator | Vitis vinifera cv. Chardonnay | [30] |
| Rice chitinase ( RCC2 ) | Oryza sativa | PR protein Class I Chitinase | Major resistance to Erysiphe necator and slight resistance to Elsinoe ampelina | Vitis vinifera cv. Neo Muscat | [31] |
| Chitinase and ribosome-inactivating protein ( RIP ) | Hordeum vulgare | These genes encode for two antifungal proteins | Susceptibility to Erysiphe necator and Plasmopara viticola infection equal to that of the control | Vitis vinifera cv. Seyval blanc | [32] |
| Rice Chitinase ( Chi 11 ) | Oryza sativa | Pathogenesis-related protein | Late and reduced manifestation of Erysiphe necator symptoms | Vitis vinifera cv. Pusa Seedless | [33] |
| Chitinase and β-1,3-glucanase | Scab-infected Sumai 3 wheat | Pathogenesis-related proteins | Chitinase was more effective than glucanase in conferring tolerance to Plasmopara viticola | Vitis vinifera cv. Crimson Seedless | [34] |
| VpPR4-1 | Vitis pseudoreticulata | PR4 proteins are considered chitin-binding proteins | Improved tolerance to Erysiphe necator | Vitis vinifera cv. Red Globe | [35] |
| Thaumatin-like protein ( Vvtl-1 ) | Vitis vinifera cv. Chardonnay | Pathogenesis-related protein 5 | Increased resistance to Erysiphe necator and Elsinoe ampelina | Vitis vinifera cv. Thompson Seedless | [36] |
|
Thaumatin-like protein
( VqTLP29 ) |
Vitis quinquangularis cv. Shang-24 | Pathogenesis-related protein 5 | Increased resistance to powdery mildew, but decreased resistance to Botrytis cinerea | Arabidopsis thaliana | [37] |
| Thaumatin-like protein ( VaTLP ) | Vitis amurensis Rupr. “Zuoshan-1” | PR5 proteins have endo-β-1,3-glucanase activity; binding β-1,3-glucan | Reinforced resistance to Plasmopara viticola | Vitis vinifera cv. Thompson Seedless | [38] |
| VpPR10.1 | Vitis pseudoreticulata | PR10 have in vitro ribonuclease activity | Increased tolerance to Plasmopara viticola | Vitis vinifera cv. Thompson Seedless | [39] |
| Antimicrobial Peptides (AMPs) | |||||
| Natural Magainin-2 (Mag2)/synthetic derivate (MS199) | Magainin extracts from the skin of Xenopus laevis frog | Magainins with broad-spectrum in vitro antimicrobial activity against bacteria and fungi | Resistance to bacterial diseases such as crown gall diseases, minor susceptibility against Erysiphe necator | Vitis vinifera cv. Chardonnay | [40] |
| Magainin-2 ( mag2 ) + PGL | Magainin extracts from the skin of Xenopus laevis frog | AMP belonging to the Magainins family | PGL protein seems to inhibit Botrytis cinerea spore germination | Vitis vinifera cv. Chardonnay | [41] |
| Transcription Factors | |||||
| VvWRKY2 | Vitis vinifera cv. Cabernet Sauvignon | WRKY protein isolated after Plasmopara viticola infection | Increased tolerance to Botrytis cinerea and broad-spectrum fungal resistance | Nicotiana tabacum cv. Xanthi | [42] |
| VvWRKY33 | Vitis vinifera | WRKY protein | Enhanced resistance to Plasmopara viticola | Vitis vinifera cv. Shiraz | [43] |
| VpWRKY3 |
Vitis pseudoreticulata accession “Baihe-35-1” |
WRKY protein isolated after Erysiphe necator infection | Improved tolerance to Ralstonia solanacearum | Nicotiana tabacum cv. NC89 | [44] |
| Ethylene response factors (VpERF2 and VpERF3) | Vitis pseudoreticulata | Transcription factor isolated after Erysiphe necator infection | Enhanced resistance to Ralstonia solanacearum and Phytophtora parasitica var. nicotianae Tucker | Nicotiana tabacum cv. NC89 | [45] |
| C-repeat-binding factor dehydration-responsive element-binding factor 1C (MrCBF2/DREB1C) | Muscadinia rotundifolia “Noble” | Transcription factor isolated after Plasmopara viticola inoculation | Enhanced resistance to Peronospora parasitica | Arabidopsis thaliana “COL0” | [46] |
| bZIP transcription factor ( VvbZIP60 ) | Vitis vinifera cv. Jing Xiu | Transcription factor that activates the accumulation of salicylic acid and the expression of PR1 protein | Enhanced resistance to powdery mildew | Arabidopsis thaliana | [47] |
| DOF protein (VvDOF3) | Vitis vinifera | Protein involved in plant growth, development, and plant defense | Enhanced resistance to powdery mildew | Arabidopsis thaliana | [48] |
| Tify protein (VvTIFY9) | Vitis vinifera | Protein highly expressed in leaves. Play an active role in SA pathway | Increased resistance to powdery mildew | Arabidopsis thaliana | [49] |
| C2H2-type zinc finger protein (VvZFP11) | Vitis vinifera | This protein expression is regulated by salicylic acid and methyl jasmonate | Enhanced resistance to powdery mildew | Arabidopsis thaliana | [50] |
| Secondary stress-related metabolites | |||||
| PR10 promot e r- Stilbene synthase (Vst1) | Vitis vinifera cv. Optima | Stilbenes production | Decreased susceptibility to Botrytis cinerea | 41B rootstock (Vitis vinifera cv. Chasselas x Vitis berlandieri) | [51] |
| Stilbene synthase (Vst1) | Vitis vinifera | Stilbenes production | Reinforced resistant against Botrytis cinerea | Vitis vinifera cv. Sugraone | [52] |
| Stilbene synthase (STS) | Vitis pseudoreticulata | Stilbenes synthesis | Transgenic plants with high resveratrol content | Vitis vinifera cv. Thompson Seedless | [53] |
| Calcium-dependent protein kinase (CDPK) (VaCPK20) | Vitis amurensis | Regulator of the biosynthetic pathways of resveratrol | Increased expression of STS7 gene, enhanced resveratrol production | Cell cultures of Vitis amurensis rupr. | [54] |
| Stilbene synthase (VpSTSgDNA2) | Vitis pseudoreticulata | Stilbenes production | Improved tolerance against Erysiphe necator | Vitis vinifera cv. Chardonnay | [55] |
| Stilbene synthase (VpSTS) | Vitis pseudoreticulata | Stilbenes production | Improved resistance to powdery mildew | Arabidopsis thaliana | [56] |
| Stilbene synthase (VaSTS19) | Vitis amurensis | Stilbenes production | Improved resistance to Botrytis cinerea and powdery mildew | Arabidopsis thaliana | [57] |
| Stilbene synthase (VqSTS6) | Vitis quinquangularis | Stilbenoids accumulation | Improved resistance to Erysiphe necator | Vitis vinifera cv. Thompson Seedless | [58] |
| Defense-related genes | |||||
| Jasmonate-ZIM domain protein (VqJAZ4) | Vitis quinquangularis clone Shang-24 | This gene is upregulated after Erisiphe necator inoculation | Improved resistance to powdery mildew and enhanced susceptibility to Botrytis cinerea. | Arabidopsis thaliana | [59] |
| Polygalacturonase-inhibiting proteins ( pPGIPs ) | Pear fruit | PGIPs are plant cell wall proteins that specifically inhibit fungal endo-polygalacturonases (PGs). | Increased resistance to Botrytis cinerea and slight tolerance to Xylella fastidiosa | Vitis vinifera cv. Thompson Seedless and Chardonnay | [60] |
| Two endochitinases (ech42 and ech33) and one N-acetyl- β -d-hexosaminidase (nag70) | Trichoderma harzianum, Trichoderma virens | Extracellular endochitinases of biocontrol agents and chitinolytic genes | Enhanced resistance to Botrytis cinerea. Tolerance to Erysiphe necator in ech42-nag70 expressing transgenic plants | Vitis vinifera cv. Thompson Seedless | [61] |
| E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (VpEIRP1) | Vitis pseudoreticulata Baihe 31-1 accession | This protein activates plant defense response through the proteolysis of VpWRKY11 transcription factor | Enhanced resistance to powdery mildew | Arabidopsis thaliana | [62] |
| F-box/Kelch-repeat protein (VpEIFP1) | Vitis pseudoreticulata | Transcription of EIFP protein is induced after powdery mildew infection and activation of PR genes | Enhanced tolerance to Erysiphe necator | Vitis vinifera cv. Red Globe and Arabidopsis thaliana | [63] |
| Metacaspases ( VrMC2 and VrMC5 ) | Vitis rupestris | Executors of hypersensitive response (HR), isolated after Plasmopara viticola infection | Programmed cell death (PCD) activation | Nicotiana tabacum cv. Bright Yellow 2 and Vitis vinifera cell cultures | [29] |
| Serotonin N-acetyltransferase ( VvSNAT2 ) | Vitis vinifera | Protein essential for melatonin production and for SA and JA signaling pathways activation | Improved resistance to powdery mildew | Arabidopsis thaliana | [64] |
| Resistance to Powdery Mildew 8 locus (RPW8.2) | Arabidopsis thaliana | Protein that encodes for small basic protein, with weak homology with NB-LRR protein | Erysiphe necator hyphal growth and sporulation were significantly restricted | Vitis vinifera cv. Thompson Seedless | [65] |
| Ubiquitin ligase ( VpPUB23 ) | Vitis pseudoreticulata | Type E3 ubiquitin ligase is involved in many immune regulation responses | Decreased resistance to Erysiphe necator | Vitis vinifera cv. Thompson Seedless | [66] |
| Ubiquitin ligase ( VaPUB ) | Vitis amurensis | U-box protein E3 ligase causes downregulation of PR10 | Transgenic plants were susceptible as control to Plasmopara viticola | Vitis vinifera cv. Thompson Seedless | [67] |
| VaHAESA | Vitis amurensis cv. Shuanghong | Pattern recognition receptor (PRR) that belongs to leucine-rich repeat receptor-like protein kinase | Induce H2O2, NO, and callose accumulation. Leaves showed less spores and Plasmopara viticola infected areas than control | Vitis vinifera cv. Thompson Seedless | [68] |
3.1. Overexpression of Pathogenesis-Related Proteins
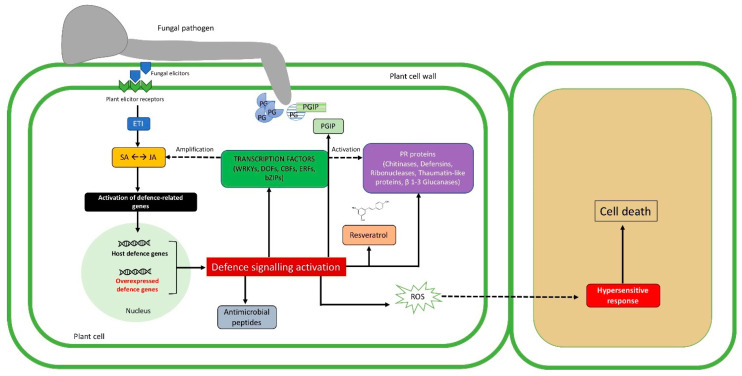
Once elicitors have been recognized by the plant, the contact with the pathogen induces different defense mechanisms in host cells, such as the reinforcement of structural barriers, the synthesis of secondary stress-related metabolites such as phytoalexins, and the provision of PR proteins [69], as depicted in Figure 1.
Figure 1.
Description of transgenic plant–pathogen arms race during pathogenesis. The first impediment to fungal invasion is represented by chemical and physical barriers already existing before the infection. The trophic activity begins with lytic enzyme production (e.g., polygalacturonase, PG), that can be suppressed by the production of specific inhibitors (e.g., polygalacturonase inhibitor, PGIP), which can be expressed also by the use of genetic engineering techniques. A specific recognition takes place when elicitors coded by avirulence (Avr) genes of the fungal cell are recognized by host receptors, driving effector-triggered immunity (ETI). ETI results in the activation of defense gene expression (i.e., defense molecules, antimicrobial peptides, phytoalexins), through the salicylic/jasmonic acid (SA/JA) signaling pathways [28]. The aforementioned defense molecules together with pathogenesis-related proteins (PRs) and transcription factors can be overexpressed in the host cell, imparting a harmful effect against the pathogen. Intriguingly, transcription factors are responsible for activating plant defense response, and their overexpression leads to the stimulation of SA/JA signaling pathways. Solid arrows and dashed arrows indicate direct or indirect induction processes, respectively.
The extreme result of ETI is the hypersensitivity response that, thanks to reactive oxygen species (ROS) accumulation, leads to programmed cell death (PCD), isolating and detaining the propagation of the pathogen in other plant cells [29].
PR proteins (PRs) are a class of soluble proteins that were isolated for the first time from tobacco tissues after infection with the tobacco mosaic virus (TMV) [70]. In general, all the proteins expressed in response to both abiotic and biotic stresses are included in this category [17]. The expression of some of these proteins can also be triggered by the accumulation of plant hormones, such as salicylic acid (SA), jasmonic acid (JA) and ethylene (ET), which are related to plant defense [17]. PRs are relevant in resistant responses against fungal attack as they are generally involved in the formation of necrotic lesions that limit pathogen invasion and growth. Furthermore, they are activated in different ways depending on the pathogen trophic behavior. Biotrophs turn on the SA pathway in the plant, that triggers NPR1 gene (non-expressor of PR gene 1) expression, which, in turn, induces the transcription and production of the SA-mediated gene proteins (PR1, PR2, PR5) that circulate in the sap, giving rise to Systemic Acquired Resistance (SAR) [17,71]. The overexpression of the Vitis vinifera NPR1.1 gene increased resistance to powdery mildew in this species through the constitutive activation of PR1 and PR2 genes expression also in uninfected plants [30]. Necrotrophs induce the activation of the JA pathway in the plant, which induces the local accumulation of JA-mediated proteins (PR4, PR5, PR12), leading to Local Acquired Resistance (LAR) [17]. At present, 17 PR families have been classified from different plant species and some of them have shown evident antifungal activity such as β 1-3 glucanases (PR2) [72], chitinases (PR3, PR4, PR8, PR11) [73,74,75], thaumatin-like proteins (PR5) [73], proteinase inhibitors (PR6) [76], peroxidases (PR9) [77], ribonuclease like-proteins (PR10) [78], defensins (PR12) [79], and thionins (PR13) [80].
Chitinases and β 1-3 glucanases show antifungal activity thanks to their direct attack on the fungi cell wall, causing its fragmentation and disaggregation [34]. Two Vitis vinifera transgenic lines, expressing the rice chitinase gene (RCC2), exhibited HR and a significant reduction in powdery mildew symptoms (suppression of both conidial germination and mycelial growth) caused by Erysiphe necator and slight resistance to anthracnose when compared with the non-transformed control [31].
Bornhoff and collaborators showed the ineffectiveness of protection against powdery and downy mildew in transgenic V. vinifera plants expressing chitinase and RIP (Ribosome-inactivating protein) isolated from barley [32], contradicting the expectation of having synergistic effect due to the coexpression of these two enzymes. On the contrary, Nookaraju and co-authors observed a low susceptibility to Plasmopara viticola, characterized by a 15 to 35% reduction in hyphal growth in plants expressing both chitinase and β 1-3 glucanase genes [34]. By using the gene construct codifying for the rice chitinase gene chi 11, researchers obtained two grapevine transgenic lines characterized by a high chitinase activity, which displayed smaller lesions and delayed manifestation of powdery mildew symptoms [33]. Many chitin-binding proteins belong to the PR4 family, described mainly as wound-inducible proteins, and triggered by fungi infection in several plants. The isolation from V. pseudoreticulata of a PR4 protein and its overexpression in the susceptible V. vinifera genotype Redglobe led to an increased resistance to powdery mildew, inhibiting hyphal growth [35].
Thaumatin proteins belong to the PR5 family and are characterized by a thaumatin domain and a PR5-like protein kinase receptor [37]. Their anti-oomycete mechanism relies on their β 1-3 glucan binding and endoglucanase activities [38]. Dhekney and colleagues obtained two cisgenic grapevine lines showing broad-spectrum antifungal resistance, by expressing a gene construct codifying for the Vitis vinifera thaumatin-like protein 1 (VVTL-1), that conferred a 10 day delay in symptoms manifestation, compared to the non-transformed control after powdery mildew infection, and a significant resistance to black rot, a fungal disease caused by Guignardia bidwellii [36]. Transgenic Vitis vinifera, expressing a thaumatin-like protein gene isolated from Vitis amurensis, showed decreased susceptibility to downy mildew, reducing the infected area and the number of sporangia [38]. Further studies in this field suggested that some thaumatin-like protein (TLP) genes, perform better against biotrophs rather than necrotrophs, like TLP29 gene of V. quinquangularis (VqTLP29) expressed in A. thaliana, where an increased susceptibility to B. cinerea was detected [37]. PR10 proteins are highly expressed after pathogen invasion, and their antifungal capability seems to also be associated with their RNase/DNase activity, and to their role in the control of flavonoid biosynthesis [81,82]. The VpPR10.1 gene inserted through Agrobacterium-mediated transformation in Thompson Seedless cultivar led to reduced hyphal growth of Plasmopara viticola, through callose deposition around hyphae and haustoria, and hydrogen peroxide accumulation compared to non-transgenic lines [39].
3.2. Gene Expression of Antimicrobial Peptides
In addition to PR proteins, scientists revealed that some antimicrobial peptides (AMPs) were proved to have antifungal activity [83]. However, overexpression of AMPs does not always result in an enhanced resistance against fungi, maybe due to the activity of endogenous proteases which can inactivate peptides, neutralizing their antimicrobial properties [84]. Magainins are a class of antimicrobial peptides that interfere with fungal membranes function, altering their polarity and inducing cell mortality, but they do not interfere with the membrane of host cells. Grapevine plants overexpressing either natural or synthetic magainins have shown enhanced resistance to Agrobacterium vitis and Erysiphe necator [40]. In another study, the ectopic expression of Magainin-2 (mag 2) and PGL genes generated plants slightly resistant to powdery mildew [41].
3.3. Overexpression of Transcriptional Factors
The role of transcriptional factors is strictly correlated with signaling pathways, and they play the role of regulating the expression of PR genes or enzymes implicated in defense responses [85]. The three more relevant transcription factor families that are unique in plants are WRKY proteins, ethylene-responsive element-binding factors (ERFs), and basic-domain leucine-zipper (bZIP) [86].
The WRKY proteins family includes 74 Arabidopsis members having the aminoacidic sequence WRKYGQK and a zinc finger-like motif in common [43]. Following Plasmopara viticola infection in grapes, VvWRKY2 gene upregulation took place before the increased expression of PR genes, suggesting its involvement in phytopathogenesis [42]. In tobacco, the ectopic expression of the VvWRKY2 gene led to a broad-spectrum resistance against fungi and oomycetes [42]. A reduction of up to 70% sporulation of P. viticola infecting grapevine leaves was also recorded in cisgenic plants expressing the VvWRKY33 gene [43], and a 40% decreased susceptibility was recorded when 41B rootstock overexpressed the VvWRKY1 gene [87].
In Arabidopsis, almost 124 ERF transcription factors are known to be involved in cold and drought tolerance as well as pathogen resistance [86]. The transcription factor VpWRKY11, that is a negative regulator of basal resistance in Arabidopsis, undergoes a proteolytic degradation operated by Erysiphe necator-induced RING finger protein 1 (EIRP1), thanks to its E3 ligase activity. Overexpression of the VpEIRP1 gene in Arabidopsis showed enhanced resistance to powdery mildew and bacterial diseases [62]. The overexpression of three ERF genes isolated from Vitis pseudoreticulata showed a different level of tolerance against pathogenic fungi in tobacco and Arabidopsis plants, suggesting that more research is required for these transcription factors [45]. Muscadinia is another genus of Vitaceae family, characterized by plants naturally immune to different pathogens, including downy mildew. The overexpression of CBF2 transcription activator isolated from M. rotundifolia in Arabidopsis gave decreased susceptibility to downy mildew (Peronospora parasitica) and enhanced cold and drought tolerance, although it caused morphological changes and flowering delay [46].
Almost 75 transcription factors with a basic region/leucine zipper motif (bZIP) have been identified in Arabidopsis, which are able to regulate different mechanisms from plant defense signaling pathways to seed and flower development [88]. The ectopic expression of bZIP60 gene, isolated from Vitis vinifera, decreased the severity of the symptoms of powdery mildew on Arabidopsis [47]. DOF transcription factors derived by the expression of the DOF gene family, which includes 25 members in Vitis vinifera, are involved in plant development and gene expression regulation [89]. The VvDOF3 gene overexpressed in Arabidopsis acted as a transcription factor and increased resistance to powdery mildew [48].
3.4. Overexpression of Secondary Metabolites Generally Induced by Biotic Stress
Grapevine cultivation became also appealing from a nutraceutical point of view, due to the abundance of molecules with high antioxidant capacity contained in grapes. Stilbene, flavonols, and anthocyanins represent the major phenolic compounds that are responsible for generating beneficial effects also to plants, and they assist plant–pathogens arms races [90]. One of the most famous compounds, known for its anticancer property, is resveratrol, a stilbene polyphenol, constitutively present at low concentration in all grapevine organs. Resveratrol tends to accumulate in areas close to Erysiphe necator, Plasmopara viticola, and Botrytis cinerea infection sites, limiting pathogen spread and diffusion since these fungi are unable to metabolize this component. The antimicrobial and antifungal activity of resveratrol has been highlighted by some studies, showing that this compound displays an inhibitory effect on Botrytis conidia germination and mycelium growth, leading to ultra-structural changes on conidia (e.g., granulation of cytoplasm and disorganization of cell content) [91].
The biosynthetic pathway common to all the major phytoalexins in grapevine requires the activity of stilbene synthase (STS), which, by condensing three Malonyl CoA molecules with one molecule of Coumaryl CoA, leads to resveratrol production [51]. In vitro infection tests with Botrytis cinerea were carried out on grapevine micro cuttings transformed with the V. vinifera Vst1 gene under the control of a promoter inducible by fungal infection (PR10 promoter isolated from alfalfa); the results obtained confirmed an enhanced resistance against the pathogen in the transgenic lines that exhibited the highest resveratrol synthesis level [51]. In another study, the expression of the Vst1 gene under the cauliflower mosaic virus 35S promoter resulted in smaller botrytis necrotic lesion size in transgenic grapevine in comparison with control plants, demonstrating the existence of a positive correlation among resveratrol content and resistance to Botrytis cinerea [52]. In transgenic Arabidopsis plants, the expression of the Vitis pseudoreticulata STS gene (VpSTS), encoding a novel stilbene synthase, led to greater resistance to powdery mildew, since its expression seemed to be stimulated by SA, as happens with the expression of its homologous allele from susceptible Vitis vinifera [56]. The same STS gene was successfully introduced in Thompson Seedless somatic embryos, showing an increased concentration of resveratrol in the transgenic lines; however, no studies on increased pathogen resistance were carried out in this work [53].
On Chardonnay transformed plants, the same target genes led to an increased production of hydrogen peroxide and consequently, reduced mycelium growth of Erysiphe necator [55]. The STS19 gene from Chinese wild grape was able to reduce powdery mildew and grey mold susceptibility when expressed in transgenic Arabidopsis plants, through SA/JA signaling pathways enhancement [57]. Although the genetic transformation aimed at improving the phytoalexin content seems to be a good strategy to reinforce plant immunity, at the same time, it can cause physiological and morphological alterations of the vine, especially during flowering and berry ripening [51].
Other molecular pathways are related to phytoalexin synthesis. It has been demonstrated that a positive correlation exists between calcium-mediated signaling and a high amount of resveratrol, corroborated by the fact that treatments with calcium channels blockers reduced resveratrol synthesis in transgenic Vitis amurensis cells [54]. The overexpression of the calcium sensor protein CDPK20 stimulated resveratrol production in Vitis amurensis cells, although only an increased VaSTS7 gene expression was elicited, while the expression of other STS genes remained unaltered [54].
3.5. Overexpression or Gene Expression of Other Defense-Related Genes
Jasmonate Zim domain (JAZ) proteins are transcriptional repressors of JA signaling pathways, and participate in secondary metabolites biosynthesis, in addition to their involvement in response to biotic and abiotic stresses [59]. VqJAZ4 gene (a jasmonate-ZIM gene from Vitis quinquangularis) transcription was induced after SA and MeJA application and by Erysiphe necator infection, evidencing its role in defense mechanisms [59]. Transgenic Arabidopsis plants expressing this gene showed enhanced resistance to powdery mildew and increased ROS accumulation and callose production, compared to non-transgenic control. However, the same VqJAZ4 expressing lines appeared more susceptible to grey mold, possibly because VqJAZ4 gene suppresses the expression of JA-related genes, and therefore, impeding JA signaling pathways activation, making the control of necrotrophs by the plant more thwarted [59].
One essential event for fungal and oomycetes pathogenesis is the production of polygalacturonases (PG) which acts in the impairment of host cell walls. The introduction of polygalacturonases inhibitors (PGIPs) in the plant through genetic engineering can limit the degradation of host cells and also as a result of enzymatic activity of cell walls compounds make available some glucosidic fragments that can react as effectors thus, eliciting host defense [60]. Twelve of the eighteen transgenic lines of Vitis vinifera expressing the pear PGIP gene showed a significant reduction in necrotic lesion size after inoculation with Botrytis cinerea [60]. In this study, the movement of PGIP through the xylem and the graft union was also demonstrated. This is an important result that shows the obtainment of genetically improved non-transgenic scion grafted on transgenic rootstock.
Trichoderma spp. are generally introduced in the field, notably in organic farming, as biocontrol agents for fungal disease management. The isolation of endochitinases and hexosaminidases from these biocontrol fungi and their genes transfer in grapevine cultivars allowed the obtainment of plants with enhanced tolerance to Botrytis cinerea and Erysiphe necator [92]. Ubiquitination in plants plays different strategic roles, especially regarding the selective degradation of proteins. The overexpression of the E3 ubiquitin ligases from Vitis pseudoreticulata led to an increased susceptibility in transgenic plants to powdery mildew [62].
Stimulation of HR represents a useful strategy, particularly effective for its rapid mode of action. It has been demonstrated that the metacaspases MC2 and MC5, identified in V. rupestris upon Plasmopara viticola inoculation, were involved in the execution of HR, activating the most efficient ETI in this genotype [29]. The ectopic expression of caspase-like regulators in plants related to cell apoptosis could increase resistance to broad-spectrum diseases, which mediates defense-related programmed cell death [29].
Melatonin is a molecule firstly discovered in mammals, where it acts as a fundamental regulator of circadian rhythms, and its presence was also confirmed in higher plants. Considering its innate antioxidant activity, this molecule can be used in agriculture both as a plant growth regulator and as a biostimulator to cope with stress conditions [93]. The melatonin biosynthetic pathway includes modifications of L-tryptophan and the acetylation of serotonin, operated by serotonin-N-acetyl transferase (SNAT). Expressing the SNAT2 gene cloned from V. vinifera in Arabidopsis plants led to a greater accumulation of melatonin and reduced susceptibility to powdery mildew [64].
In the Arabidopsis genome, a locus that confers resistance to different mildews, RPW8, has been characterized and it is composed of two genes, RPW8.1 and RPW8.2 [94]; the latter is located in the extrahaustorial membrane that covers the fungal haustorium and promotes accumulation of hydrogen peroxide. The expression of this gene (characterized by an efficient inhibition of fungal growth and sporulation) in transgenic Vitis vinifera plants demonstrated that this resistance can be transferred in this species as well [65].
PTI in plants could be promoted by the expression of PRRs dedicated to PAMPs recognition. Transient expression of VaHAESA, a V. amurensis leucine-rich repeat receptor-like protein kinase, in grapevine leaves, determined significant reduction in an infected area by downy mildew, assisted by callose deposition, H2O2, and NO accumulation on the nearby infection site [68].
4. RNAi: Host- or Spray-Induced Gene Silencing against Fungi and Oomycetes
In addition to owning genetic heritage, nucleic acid molecules can be managed as tools that follow specific recognition by the vegetal cell and can be processed in various ways, giving a start to a set of signals that may stimulate the induction of different defense responses up to the block of specific mRNA translation [95].
Post-transcriptional gene silencing is arbitrated by the activity of small RNAs (sRNAs) [96]. In the plant kingdom, the class of sRNAs is represented by microRNAs (miRNAs) originating from endogenous MIR loci, and small interfering RNAs (siRNAs) derived from long double-stranded RNA (dsRNA) molecules [97]. Using RNA templates, RNA-dependent RNA polymerases (RDRP) are responsible for de novo synthesis of secondary siRNAs that exert a stronger role in gene silencing than primary siRNAs [98]. This eukaryotic conserved silencing mechanism allows transcripts degradation or protein production restraint, through sRNAs production having full or partial complementary sequences with the target mRNA. sRNAs are loaded into argonaute (AGO) protein, part of RISC complex (RNA-induced silencing complex), where they become probes for binding with complementary RNA targets, thus, exerting their silencing ability [20].
Host defense mechanisms and pathogen virulence strategies are linked through cross-kingdom mechanisms [96]. In the same way the plant sends siRNAs to silence target genes of the pathogens, the pathogen uses the same mechanism to increase its virulence by impairing host immunity genes [97,99]. Bidirectional cross-kingdom RNAi can be exploited for generating silencing effects through the introduction of RNA molecules in transgenic plants that can counteract fungal and oomycetes virulence genes [100]. Nowara and collaborators showed that the accumulation of dsRNAs in barley (Hordeum vulgare) and wheat (Triticum aestivum) targeting the fungal glucanosyl transferase, Avrk1 and Avra10 effectors caused a reduction in haustorium formation of the causative agent of powdery mildew Blumeria graminis [101]. Gene silencing of other effector proteins led to similar results against powdery mildew in Hordeum vulgare [102,103].
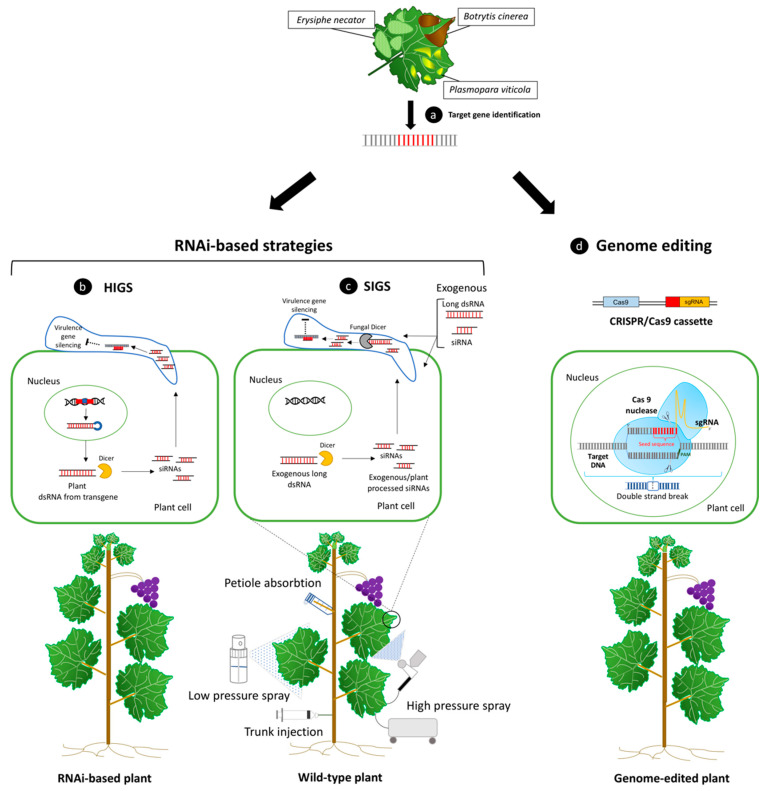
Once one or more genes to be silenced have been identified by studying plant–pathogen interaction processes (Figure 2a), several approaches can be used to deliver dsRNAs into plants. The potential of a host-induced gene silencing (HIGS) approach in crop disease management can be explored through the expression of RNAi constructs against various target genes in the host, as shown in Figure 2b. Resistance to Fusarium graminaerum was obtained by HIGS approach in Arabidopsis and barley plants, by targeting the fungal sterol 14α demethylase (CYP51) [104]. Other Fusarium spp. related diseases were successfully controlled by HIGS strategy in embryogenic cells of banana targeting two FOC proteins, whose roles are strongly related to fungal growth, development, and pathogenesis [105] or chitinase genes in Triticum aestivum [106]. Barley-stripe mosaic virus-induced RNAi worked efficiently also against Puccinia tritici, hitting three pathogenicity genes, and reducing leaf wheat rust [107]. Downregulation of the Verticillium dahliae hygrophobins 1 (VdH1) gene, important for microsclerotia production, led to reduced wilt symptoms in cotton transformed plants [108].
Figure 2.
In addition to trans/cisgenesis methods, the expression of RNAi gene constructs in the plant, the exogenous applications of double strand RNA (dsRNA) molecules targeting host/pathogen genes, or plant genome editing, represent valid alternatives to enhance plant immunity during pathogenesis. (a) Candidate genes capable of limiting pathogen aggression or improve plant defense responses can be identified during the infection processes caused by the fungal and oomycetes causal agents of the most impactful diseases for grapevine production. RNAi-based strategies can be exploited to improve plant defense by providing dsRNAs to the plant cell through the expression of an introgressed hairpin-based gene construct in the plant genome, or through their delivery by exogenous application. (b) In host-induced gene silencing (HIGS), as a result of transcription of an RNAi sequence, a long dsRNA molecule is formed. When this molecule is recognized by Dicer-like protein, it is cleaved into siRNAs, which can knockdown related target gene expression [100]. (c) A transgenic-free procedure in which dsRNAs are directly sprayed on the surface of plants and pathogens is known as spray-induced gene silencing (SIGS). These molecules can be absorbed by both types of cells, and, depending on the delivery method used, dsRNAs can be processed by either the fungal/oomycetes and host RNAi machinery, leading to virulence gene knockdown and reduction in pathogen detrimental effects. Low-pressure spray, high-pressure spray, petiole adsorption, and trunk injection of dsRNAs represent some of the different available exogenous dsRNA delivery methods to confer plant protection against different plant pathogens, included fungi [143]. d) CRISPR/Cas9 system can be used for inducing targeted genome editing in plants, including the inactivation of specific plant susceptibility genes expression, which can help to regulate plant–pathogen interaction processes and disease resistance enhancement. Cas 9 protein complex is guided by artificially designed single guide RNA molecule (sgRNA) and leads to double-strand breaks (DSBs) of targeted DNA. SgRNA contains a seed sequence (around 8–12 bp, shown in red) complementary to target DNA that guides the binding of the Cas 9 protein to the target genomic sequence. The site of cleavage takes place three nucleotides upstream to the protospacer adjacent motif (PAM, shown in green) [20].
RNAi machinery has been demonstrated to be also functional against oomycetes pathogens. Significant reduction in Phytophtora spp. load and disease progression was recorded in HIGS potato plants targeting the Avr3a effector, and the G protein-β-subunit 1 (PiGPB1) gene of this pathogen species [109,110,111].
HIGS strategy has also been applied in lettuce plants to express silencing constructs targeting Highly Abundant Message (HAM34) or Cellulose Synthase (CES1) genes of Bremia lactucae, making these plants resistant to downy mildew [112].
All these studies have demonstrated the efficacy of this technique (HIGS) in specific genes downregulation, highlighting their high potential that can be managed in future grapevine genetic improvement programs aimed at increasing resistance to biotic stresses.
In addition to gene silencing against an external pathogen, is it also possible to exploit the RNAi mechanism to target endogenous genes, that have a negative influence in the pathosystem. An example is the case of MLO (Mildews Locus O) genes with transmembrane domain, considered as susceptibility genes (S-genes), that alter vesicle-associated and actin-dependent defense pathways [113]. Knockdown of the VvMLO7 gene through constitutive expression of long non-coding dsRNA led to a significant reduction in powdery mildew disease severity in the transgenic grapevine cultivar Brachetto [114].
However, the application of the HIGS approach is limited by poor public acceptance and strict legislative rules applied to GMO cultivation, and also by the lack of efficient in vitro regeneration and genetic transformation protocols for several crops, and more often, for all the genotypes within the same species [115].
The fact that simple exogenous application of polynucleotides can affect mRNA levels of important virulence-related genes of pathogens/plants without modifying the host genome, opens new opportunities for the development of new scientific techniques and crop improvement strategies [116]. Extracellular-self DNA and RNA could be also applied to the plants in order to stimulate their immune response [97]. The attack of siRNA production machinery in the pathogen, through RNAi, has the potential to inhibit the pathogen virulence itself. Wang and collaborators demonstrated that Botrytis cinerea DCL1/2-long dsRNAs, targeting expression of dicer proteins essential for sRNA production, exogenously applied on the surface of detached leaves and fruits of different plant species, including grapes, can be efficiently taken up by the necrotrophic fungus, providing a relevant protection against grey mold [100]. Similar observations were made when spray applications of long non-coding dsRNA molecules, which target three genes required for the biosynthesis of Fusarium graminearum ergosterol, efficiently inhibited the fungal growth at the sprayed (local) as well as the non-sprayed (distal) parts of detached leaves, probably due to the basipetal and acropetal transportation along the vascular system of the silencing signal [117]. In many cases, researchers preferred to adopt a multitarget approach, by silencing simultaneously two or more target genes entailing in pathogenesis. White mold and grey mold symptoms can be significantly decreased in Arabidopsis and Brassica napus leaves, respectively, through foliar application of 20 different dsRNAs targeting various genes, evidencing the possibility to counteract closely related fungi while applying the same dsRNAs molecules on various crops [118]. The same conserved target gene among various fungi, such as β2-tubulin gene of Fusarium asiaticum, could be selected for RNAi, altering the damaging effects afforded by Fusarium spp., Botrytis cinerea, Magnaporthe oryzae, and Colletotrichum truncatum [119].
In addition to HIGS, the exogenous application of long dsRNAs, small dsRNAs, and hairpin RNAs has been recently studied and proposed as a new environment-friendly crop protection tool [120]. Spray-induced gene silencing (SIGS) allows the adsorption of dsRNA by either plant cells and tissues, where it can be processed from host RNAi machinery and/or then progressively conveyed on pathogen cells, or directly adsorbed and processed by the fungal cell driving gene silencing through their own RNAi machinery (Figure 2c) [117,121]. However, the exact mechanisms behind the uptake of exogenous dsRNAs and their use to activate RNAi machinery in the plant and/or pathogen cells is still unclear, and they seem to be affected by the method of exogenous application used combined with the absorption capacity of different plant organs [122,123,124,125].
The appearance of pathogens-resistant strains to fungicide can be counteracted using fungicides with different modes of action or with a combined application of dsRNAs. A reduction in Fusarium asiaticum pathogenicity and resistance to phenamacril, caused by a mutation in the myosin-5-gene (Myo-5), was recorded with the continuous application of the phenamacril and dsRNA-Myo-5 as treatments on wheat speaklets [126]. Long or small dsRNAs could be supplied to plants via low-pressure or high-pressure spray, petiole adsorption, or trunk injection [98,100,117,118]. Through petiole adsorption and trunk injection methods, dsRNAs were shown to be limited to the apoplast and transported only along the xylem, without penetrating the plant cell. These results were demonstrated by Dalakouras and co-authors that applied both these techniques into Vitis vinifera, observing that the delivered hairpin-RNAs (hpRNAs) were systemically transported and detected in leaves, distant from the treated area, from one up to 10 days post-application, but no siRNAs deriving from DCL-processed hpRNAs were found [122]. Furthermore, when siRNAs were applied by petiole absorption also into GFP-expressing N. bethamiana, no silencing effect on GFP transcripts was observed [122]. Nevertheless, it seems possible by using these exogenous application techniques to directly reach fungal or oomycetes that normally colonize the apoplast and xylematic tissue where, after undergoing internal processing, dsRNAs can exert their biological activity [98]. On the other hand, high-pressure spray of siRNAs had the potential to ensure both local and systemic gene silencing on tobacco plants [127].
Selecting fungicide sites of action, Nerva and colleagues constructed a single long dsRNA molecule that exerted protection against grey mold in vitro on grapevine detached leaves and grapes at post-harvest, applied through the high-pressure spray and petiole adsorption. Despite different levels of protection being recorded among the dsRNAs delivery methods, interestingly, all the techniques that facilitate the provision of intact dsRNA to the fungus were assumed as effective [128].
Some authors have also reported the possibility to use SIGS to target endogenous genes in plants and downregulate their mRNA levels both locally and systemically [116,127,129]. To our knowledge, the data present in the literature are limited to the foliar application of dsRNA molecules to silence transgenes expressed in model plant species, like Arabidopsis and N. benthamiana; however, these results open new scenarios for the use of SIGS also to target endogenous gene sequences, like susceptibility genes in grapevine and other crops, to enhance plant defense responses.
5. Genome Editing
Genome editing is a powerful technique that facilitates the generation of multiple types of genome modifications, like insertion, deletion, or mutation, having various implications in genetic studies of animal and vegetal cells [20].
Among the three typologies of engineered nucleases that are at the base of genome editing techniques, Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) (CRISPR/Cas9) is a lower cost, simpler, and faster system compared with the other enzymes such as zinc finger nuclease (ZFN) or transcription activator-like effector nuclease (TALEN) [130]. Specific modifications in DNA sequence could be driven by the CRISPR/Cas9 reprogrammed system, which needs the insertion of a well-designed single guide RNA (sgRNA) molecule into cells through different types of vectors (Figure 2d) [130]. SgRNAs are constituted by a single molecule of RNA composed of specific crRNA-trascrRNA (transactivating RNA) chimera sequence [20]. Mutation efficiency could be notably increased by designing multiple target sgRNAs for one target gene [131]. Ren and collaborators introduced a single plasmid containing specific sgRNA through Agrobacterium-mediated transformation into Chardonnay suspension cells to alter the biosynthetic pathway of tartaric acid [132]. L-idonate dehydrogenase (idnDH) gene was successfully mutated using CRISPR/Cas9 system, without recording off-target events and highlighting the importance of high GC content in sgRNA sequence in order to obtain high efficiency in genome modifications [132]. In fact, previous research works carried out in other plant species showed that sgRNAs designed to have a GC content above 50% led to a higher editing efficiency; this might have been due to the final binding capability of these molecules to their targets, which, in some species, genomes have high GC contents in specific regions [133,134]. Vitis vinifera Phytoene Desaturases (VvPDS) was efficiently knockout in cell masses of Neo Muscat [135], Chardonnay, and 41B rootstock by CRISPR/Cas9 binary vectors expressing a sgRNA with 65% GC content [136]. Notably, the first application of target genome editing (TGE) for increasing resistance against B. cinerea in grapevine was reported by Wang and colleagues [131]. The mutations of the VvWRKY52 gene, induced by TGE in Thompson Seedless transgenic plants, led to a significant reduction in B. cinerea colonies, especially in biallelic grapevine mutant lines [131]. Mutation efficiency driven by the CRISPR/Cas9 system is widely dependent on different factors (technical methods, plant genotype, gene target, in vitro regeneration, and selective conditions) as already known for genetic transformation techniques, and others specifically for this approach such as the choice of Cas9 promoter and sgRNA sequence [137]. As an alternative to classical genetic transformation, a plasmid-mediated procedure that can lead to the generation of transgenic-free new varieties, which is based on direct delivery of CRISPR/Cas9 ribonucleoproteins (RNPs) generally into protoplasts, is available [138]. Although in vitro plant regeneration of protoplast can be applied to some herbaceous species, in recalcitrant woody fruit plants species, the development of this technique is hampered by many factors, and attempts on grapevine are relatively recent and need further studies [138,139].
A plasmid-free method to obtain genome-edited plants was elaborated by Malnoy and collaborators, in which CRISPR/Cas9 RNPs were directly inserted in protoplasts of grapevine cultivar Chardonnay and apple cultivar Golden delicious. Grapevine protoplasts were obtained from embryogenic calli, and the induction of site-directed mutation of the Mildew Locus O-7 (MLO7) gene was demonstrated, however, the regeneration of new genome-edited plants was not reported [140]. Direct delivery of CRISPR/Cas9 RNPs was also described by Osakabe and colleagues, who detailed the advantages and critical steps in the obtainment of mutated IdnDH grapevine plants regenerating from protoplast or directly regenerated after classical Agrobacterium-mediated transformation [138]. In whichever manner, genome editing technology could be effectively applied for grapevine susceptibility gene knockout, which would be a beneficial plant defense strategy. This is the case of some Vitis vinifera cultivars, where susceptibility against downy and powdery mildews was decreased through the exploitation of the CRISPR/Cas9 system, by transforming embryogenic calluses in order to induce target mutagenesis of specific susceptibility genes [141]. In the future, targeted genome editing can be exploited to insert new genes or modify genes regulating plant–pathogen interaction at the expense of pathogens. Different to other NBTs that are based on the introduction of foreign DNA sequences in the host genome, TGE represents an innovative method that can induce specific modification in the existing genome limited to the introduction of single-point mutations [142].
6. Biosafety Considerations and Overview of Breeding Technologies Applied to Enhance Resistance against Fungal and Oomycetes Disease in Grapevine
Nowadays, various methods for grapevine genetic improvement are available, including both traditional breeding methods and new biotechnological approaches. The development and application of each of these strategies is often linked to several technical advantages and disadvantages; furthermore, they often give rise to new biosafety issues and public concerns (Table 2). A brief description of the new breeding techniques, compared with traditional breeding systems, referring to the possibility of increasing in grapevine resistance to fungal and oomycetes diseases, is reported below.
Table 2.
Description of different biotechnological approaches highlighting biosafety concerns and consumer acceptance.
| Technology | Type of Modification | Target Origin and Description | Time Needed | Classification According to EU-Legislation | Side Effects | Biosafety Concerns | Consumer Acceptance (Proposed) |
|---|---|---|---|---|---|---|---|
| Traditional breeding | Breeding and several backcrosses generation, introgression breeding, induced mutagenesis, and somatic hybridization | Genes found in crossable, sexually compatible organisms | At least 10-15 years | Non-GMO | Altered clone identity, partial resistance to biotic stresses | No biosafety concerns and basic regulation needed [149] | High [153] |
| Transgenesis | Genetic transformation | Overexpression of genes also from non-sexually compatible organisms, presence of gene sequences (i.e., promoter, selectable marker gene) from non-compatible organisms | Around 1 or 2 years | GMO | Release in the environment of genes of different origins; expression of new protein products with possible allergen/toxic effects | Expression of unknown protein/enzyme; use of antibiotic/herbicide resistance markers, lack of coexistence with non-GM, organic cultivations | Low [147,154,155,156,157] |
| Cisgenesis/Intragenesis | Genetic transformation | Expression/overexpression of a gene originating from the recipient plant itself (cisgenesis), or expression of full/partial coding sequence originating from a sexually compatible plant (intragenesis) | Around 1 or 2 years | GMO | Scarce availability of efficient cisgenic selectable marker genes | cisgenic/intragenic plants solve the current biosafety problems regarding the presence of foreign genes in the plant host genome [158] | Medium/High [148,149,159] |
| Gene silencing-HIGS | Genetic transformation | Overexpression of non-coding dsRNAs downregulating exogenous or endogenous gene expression | Around 1 or 2 years | GMO | Efficacy of gene silencing varies with the genes and target organisms. Possible off-target effects in non-target organisms (NTOs) |
Reduced off-target effects by designing RNAi sequences with high specificity and verified with bioinformatic studies. Minimal biosafety concerns when HIGS is applied only to rootstocks by trans-grafting technique [160] | Medium [161,162,163] |
| Gene silencing-SIGS | No genetic modification | External application of non-coding dsRNAs downregulating exogenous or endogenous gene expression | Few months | Non-GMO | Efficacy of gene silencing depends on the efficiency and specificity of the RNAi sequence and on the degree of adsorption showed by plants and pathogens cells | RNAi sequence should be selected in order to avoid off-target effects [143]. The absence of negative effects, that can be caused by the nanotechnology-based delivery method used, on the environment and human health needs to be demonstrated | Medium/High [21,164] |
| Genome editing | Genetic transformation/plasmid-free protoplast transformation | Artificially gene modification/target random mutation | About 1 or 2 years. More time necessary if transgene segregation is required from T0 plants, or if protoplast in vitro regeneration is required | Non-GMO/GMO in Europe (ECJ-2018) |
Possible appearance of off-target mutations; difficulties in plant regeneration from protoplasts | Transgene integration, effect of the expression of Cas9 protein, specificity and fidelity of Cas9 protein [165] | Medium/High [166,167] |
Traditional breeding: With the application of this technology, it must be accepted that new cultivars will be similar to the original clones but not the same, evidencing the importance of assisted tools such as Marker Assisted Selection (MAS) that, thanks to the possibility of using molecular markers, can be employed to detect genes of interest [144]. Cultivars suitability for the wine market must be tested and classified as a new type of wine. Generally, this is a long-term program, that requires deep knowledge of genetic resources, and the new clones corresponding to the original clones recognized internationally for particular wine brands can be identified with difficulty [145,146]. Moreover, if resistance to diseases is provided by the insertion of R genes from less susceptible genotypes, it will be easily overcome by the onset of new pathogen strains [145].
Transgenesis: this technique allows the overexpression of both homologous and heterologous genes, including antifungal proteins. Whenever regeneration/transformation protocols are available, this technology can confer stable resistance to diseases to any grapevine cultivars, mostly preserving the agronomic characteristics of the original clone. Great potential is given by the high availability of gene vectors and selectable marker genes. The presence of transgenes in the plant genome and its release into the environment make risk assessment and public acceptance more difficult [147].
Cisgenesis/Intragenesis: these technologies make it possible to introduce genes originally present in the same species or in sexually compatible ones into one genotype, through genetic transformation [148,149,150]. Compared to transgenesis, despite the availability of several grapevine resistance genes, it is more difficult to create full cisgenic gene constructs due to the lack of efficient cisgenic promoters and selectable markers, thus, making the selection of stably transformed plants more complicated [20] (Table 2).
Gene silencing—HIGS-RNAi technology: HIGS uses the same transgenesis approach but the inserted RNAi gene construct can be designed with high specificity and minimization of off-target effects [20]. It is applicable to downregulate/modulate the expression of plant endogenous genes and to target genes of grapevine pests and diseases. The expression of a new short RNAi sequence instead of new proteins facilitates risk assessment [151]. With the increasing knowledge of pests, fungi, and virus genomes, this technology offers an effective and flexible tool for introducing stable resistances in grapevine cultivars.
Gene silencing—SIGS-RNAi technology: This is not considered a transgenic approach because it is not based on recombinant DNA technology, and it involves the application of small RNA molecules, with a much higher target effect. The new products are regulated as new natural molecules and not as GMO. To ensure better delivery of dsRNAs, new formulates and production systems that will reduce production costs are under validation [21]. It has been demonstrated that the SIGS pathway is greatly independent of the canonical defense pathways, hence, conferring a “less expensive” and efficient immunity to cells, in comparison to an active pattern- or effector-triggered immunity (PTI/ETI) in progress, which is expensive in terms of cellular energy [117].
Genome editing—CRISP/Cas9: this is the most recent technology for inducing target mutations in grapevine. The potential of this technology applied to grapevine depends on the identification of specific susceptibility gene sequences to be modified in the grapevine genome. Some important results have already been identified; however, the results can be affected by the type of target gene [114,132,138,140]. Different studies have demonstrated the risk of this technology in inducing off-target effects, even though it has reduced risk on the environment and on the consumer [152]. The efficiency of this technology also depends on the methods used for the insertion of CRISPR/Cas9 protein needed for genome editing. In the case of genome editing induced by genetic transformation, the new plants are definitely GMO and regulated as such. The limited availability of efficient regeneration protocols from somatic tissue or protoplasts remains the main limiting factor in applying this technology for targeting mutation in different grapevine cultivars.
7. Conclusions
Plants and pathogenic fungi/oomycetes are living organisms and their interaction give rise to a series of interlocking events culminating in plants immunity deficiency or vulnerability. The establishment of a plant disease is a complex mechanism whose resolution can be achieved by the application of integrating defense strategies. Gene overexpression, gene silencing, and genome editing are mainly used for studying gene functions and can be efficiently exploited to control pathogenic diseases caused by the Fungi and Chromista kingdoms. All these biotechnological approaches could be exploited for decreasing both pathogens’ virulence and plant susceptibility to diseases. In contrast to traditional breeding methods, the application of biotechnological techniques allows the breeder to act specifically at the gene level, avoiding the introduction of undesirable genes in the new improved grapevine cultivar. A detailed study of candidate genes involved during the infection process is required in order to select the best target for protecting plants or counteracting pathogenicity and virulence gene expression.
Regeneration and transformation of recalcitrant Vitis vinifera cultivars remains to be the biggest challenge for the application of genetic engineering-related biotechnologies [115,132]. Once engineered plants have been obtained and before their commercialization, they must be subjected to strict regulation in order to guarantee the safety of their products towards the environment and on humans thereof. For this reason, researchers started to develop alternative strategies to classical biotechnological tools such as SIGS or cisgenesis and intragenesis, avoiding the introgression in the host genome of foreign genes and the use of antibiotic resistance genes as selectable markers [115].
Altogether, these approaches have the opportunity to offer preservation of plant health during the pathogenic challenge by providing a broad spectrum of defense mechanisms, ranging from an overproduction of various compounds to RNA-mediated silencing, passing through specific gene inactivation.
Author Contributions
Conceptualization, S.S., B.M. and E.B.; writing—original draft preparation, L.C.; writing—review and editing, S.S., B.M, C.L. and E.B.; visualization, S.S., L.C..; supervision, S.S., E.B., C.L. and B.M.; project administration, B.M.; funding acquisition, B.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by Ampelos, Italian viticulture nursery consortium.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Fournier-Level A., Lacombe T., Le Cunff L., Boursiquot J.M., This P. Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.) Heredity. 2010;104:351–362. doi: 10.1038/hdy.2009.148. [DOI] [PubMed] [Google Scholar]
- 2.Vezzulli S., Zulini L., Stefanini M. Genetics-assisted breeding for downy/powdery mildew and phylloxera resistance at fem. BIO Web Conf. 2019;12:01020. doi: 10.1051/bioconf/20191201020. [DOI] [Google Scholar]
- 3.De la Fuente Lloreda M. Use of hybrids in viticulture. A challenge for the OIV. OENO One. 2018;52:231–234. doi: 10.20870/oeno-one.2018.52.3.2312. [DOI] [Google Scholar]
- 4.Real A.C., Borges J., Cabral J.S., Jones G.V. Partitioning the grapevine growing season in the Douro Valley of Portugal: Accumulated heat better than calendar dates. Int. J. Biometeorol. 2015;59:1045–1059. doi: 10.1007/s00484-014-0918-1. [DOI] [PubMed] [Google Scholar]
- 5.Armijo G., Espinoza C., Loyola R., Restovic F., Santibáñez C., Schlechter R., Agurto M., Arce-Johnson P. Grape and Wine Biotechnology. IntechOpen; Rijeka, Croatia: 2016. Grapevine Biotechnology: Molecular Approaches Underlying Abiotic and Biotic Stress Responses. [DOI] [Google Scholar]
- 6.Bois B., Zito S., Calonnec A. Climate vs grapevine pests and diseases worldwide: The first results of a global survey. OENO One. 2017;51:133–139. doi: 10.20870/oeno-one.2017.51.2.1780. [DOI] [Google Scholar]
- 7.Cortiñas Rodríguez J.A., González-Fernández E., Fernández-González M., Vázquez-Ruiz R.A., Aira M.J. Fungal Diseases in Two North-West Spain Vineyards: Relationship with Meteorological Conditions and Predictive Aerobiological Model. Agronomy. 2020;10:219. doi: 10.3390/agronomy10020219. [DOI] [Google Scholar]
- 8.Boso S., Gago P., Santiago J.L., La Fuente M.D., Martínez M.C. Factors affecting the vineyard populational diversity of Plasmopara viticola. Plant Pathol. J. 2019;35:125–136. doi: 10.5423/PPJ.OA.09.2018.0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Bracero M., Alcázar P., Velasco-Jiménez M.J., Galán C. Fungal spores affecting vineyards in Montilla-Moriles Southern Spain. Eur. J. Plant Pathol. 2019;153:1–13. doi: 10.1007/s10658-018-1532-6. [DOI] [Google Scholar]
- 10.Toffolatti S.L., De Lorenzis G., Costa A., Maddalena G., Passera A., Bonza M.C., Pindo M., Stefani E., Cestaro A., Casati P., et al. Unique resistance traits against downy mildew from the center of origin of grapevine (Vitis vinifera) Sci. Rep. 2018;8:12523. doi: 10.1038/s41598-018-30413-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalla Costa L., Malnoy M., Gribaudo I. Breeding next generation tree fruits: Technical and legal challenges. Hortic. Res. 2017;4:17067. doi: 10.1038/hortres.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guimier S., Delmotte F., Miclot A.S., Fabre F., Mazet I., Couture C., Schneider C., Delière L. OSCAR, a national observatory to support the durable deployment of disease-resistant grapevine cultivars. Acta Hortic. 2019;1248:21–33. doi: 10.17660/ActaHortic.2019.1248.4. [DOI] [Google Scholar]
- 13.Hussain S., Siddique T., Saleem M., Arshad M., Khalid A. Chapter 5 Impact of Pesticides on Soil Microbial Diversity, Enzymes, and Biochemical Reactions. Adv. Agron. 2009;102:159–200. doi: 10.1016/S0065-2113(09)01005-0. [DOI] [Google Scholar]
- 14.Parmar N., Singh K.H., Sharma D., Singh L., Kumar P., Nanjundan J., Khan Y.J., Chauhan D.K., Thakur A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech. 2017;7:239. doi: 10.1007/s13205-017-0870-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing H., Lawrence C.B., Chambers O., Davies H.M., Everett N.P., Li Q.Q. Increased pathogen resistance and yield in transgenic plants expressing combinations of the modified antimicrobial peptides based on indolicidin and magainin. Planta. 2006;223:1024–1032. doi: 10.1007/s00425-005-0143-6. [DOI] [PubMed] [Google Scholar]
- 16.Wally O., Punja Z.K. Genetic engineering for increasing fungal and bacterial disease resistance in crop plants. GM Crop. 2010;1:199–206. doi: 10.4161/gmcr.1.4.13225. [DOI] [PubMed] [Google Scholar]
- 17.Ali S., Ganai B.A., Kamili A.N., Bhat A.A., Mir Z.A., Bhat J.A., Tyagi A., Islam S.T., Mushtaq M., Yadav P., et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018;212–213:29–37. doi: 10.1016/j.micres.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Gentile A., La Malfa S. Innovations in Sustainable Agriculture. Springer International Publishing; Cham, Germany: 2019. New Breeding Techniques for Sustainable Agriculture; pp. 411–437. [DOI] [Google Scholar]
- 19.Holme I.B., Wendt T., Holm P.B. Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 2013;11:395–407. doi: 10.1111/pbi.12055. [DOI] [PubMed] [Google Scholar]
- 20.Limera C., Sabbadini S., Sweet J.B., Mezzetti B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017;8:1418. doi: 10.3389/fpls.2017.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taning C.N., Arpaia S., Christiaens O., Dietz-Pfeilstetter A., Jones H., Mezzetti B., Sabbadini S., Sorteberg H., Sweet J., Ventura V., et al. RNA-based biocontrol compounds: Current status and perspectives to reach the market. Pest Manag. Sci. 2020;76:841–845. doi: 10.1002/ps.5686. [DOI] [PubMed] [Google Scholar]
- 22.Kortekamp A., Zyprian E. Leaf Hairs as a Basic Protective Barrier against Downy Mildew of Grape. J. Phytopathol. 1999;147:453–459. doi: 10.1111/j.1439-0434.1999.tb03850.x. [DOI] [Google Scholar]
- 23.Voigt C.A. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 2014;5:168. doi: 10.3389/fpls.2014.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellinger D., Naumann M., Falter C., Zwikowics C., Jamrow T., Manisseri C., Somerville S.C., Voigt C.A. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 2013;161:1433–1444. doi: 10.1104/pp.112.211011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 26.Newman M.A., Sundelin T., Nielsen J.T., Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boutrot F., Zipfel C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- 28.Poltronieri P., Marrazzo M.T., Cipriani G. Applied Plant Biotechnology for Improving Resistance to Biotic Stress. Elsevier Inc.; Amsterdam, The Netherlands: 2020. Grapevine: Resistance genes, sRNAs and immunity; pp. 151–179. [DOI] [Google Scholar]
- 29.Gong P., Riemann M., Dong D., Stoeffler N., Gross B., Markel A., Nick P. Two grapevine metacaspase genes mediate ETI-like cell death in grapevine defence against infection of Plasmopara viticola. Protoplasma. 2019;256:951–969. doi: 10.1007/s00709-019-01353-7. [DOI] [PubMed] [Google Scholar]
- 30.Le Henanff G., Farine S., Kieffer-Mazet F., Miclot A.S., Heitz T., Mestre P., Bertsch C., Chong J. Vitis vinifera VvNPR1.1 is the functional ortholog of AtNPR1 and its overexpression in grapevine triggers constitutive activation of PR genes and enhanced resistance to powdery mildew. Planta. 2011;234:405–417. doi: 10.1007/s00425-011-1412-1. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto T., Iketani H., Ieki H., Nishizawa Y., Notsuka K., Hibi T., Hayashi T., Matsuta N. Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens. Plant Cell Rep. 2000;19:639–646. doi: 10.1007/s002999900174. [DOI] [PubMed] [Google Scholar]
- 32.Bornhoff B.A., Harst M., Zyprian E., Töpfer R. Transgenic plants of Vitis vinifera cv. Seyval blanc. Plant Cell Rep. 2005;24:433–438. doi: 10.1007/s00299-005-0959-3. [DOI] [PubMed] [Google Scholar]
- 33.Nirala N.K., Das D.K., Srivastava P.S., Sopory S.K., Upadhyaya K.C. Expression of a rice chitinase gene enhances antifungal potential in transgenic grapevine (Vitis vinifera L.) Vitis. 2010;49:181–187. [Google Scholar]
- 34.Nookaraju A., Agrawal D.C. Enhanced tolerance of transgenic grapevines expressing chitinase and β-1,3-glucanase genes to downy mildew. Plant Cell Tissue Organ Cult. 2012;111:15–28. doi: 10.1007/s11240-012-0166-1. [DOI] [Google Scholar]
- 35.Dai L., Wang D., Xie X., Zhang C., Wang X., Xu Y., Wang Y., Zhang J. The Novel Gene VpPR4-1 from Vitis pseudoreticulata Increases Powdery Mildew Resistance in Transgenic Vitis vinifera L. Front. Plant Sci. 2016;7:695. doi: 10.3389/fpls.2016.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhekney S.A., Li Z.T., Gray D.J. Grapevines engineered to express cisgenic Vitis vinifera thaumatin-like protein exhibit fungal disease resistance. In Vitro Cell. Dev. Biol.-Plant. 2011;47:458–466. doi: 10.1007/s11627-011-9358-3. [DOI] [Google Scholar]
- 37.Yan X., Qiao H., Zhang X., Guo C., Wang M., Wang Y., Wang X. Analysis of the grape (Vitis vinifera L.) thaumatin-like protein (TLP) gene family and demonstration that TLP29 contributes to disease resistance. Sci. Rep. 2017;7:4269. doi: 10.1038/s41598-017-04105-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He R., Wu J., Zhang Y., Agüero C.B., Li X., Liu S., Wang C., Walker M.A., Lu J. Overexpression of a thaumatin-like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine. Protoplasma. 2017;254:1579–1589. doi: 10.1007/s00709-016-1047-y. [DOI] [PubMed] [Google Scholar]
- 39.Su H., Jiao Y.T., Wang F.F., Liu Y.E., Niu W.L., Liu G.T., Xu Y. Overexpression of VpPR10.1 by an efficient transformation method enhances downy mildew resistance in V. vinifera. Plant Cell Rep. 2018;37:819–832. doi: 10.1007/s00299-018-2271-z. [DOI] [PubMed] [Google Scholar]
- 40.Vidal J.J., Kikkert J.R., Malnoy M.A., Wallace P.G., Barnard J., Reisch B.I. Evaluation of transgenic “Chardonnay” (Vitis vinifera) containing magainin genes for resistance to crown gall and powdery mildew. Transgenic Res. 2006;15:69–82. doi: 10.1007/s11248-005-4423-5. [DOI] [PubMed] [Google Scholar]
- 41.Rosenfield C.-L., Samuelian S., Vidal J.R., Reisch B.I. Transgenic Disease Resistance in Vitis vinifera: Potential Use and Screening of Antimicrobial Peptides. Am. J. Enol. Vitic. 2010;61:348–357. [Google Scholar]
- 42.Mzid R., Marchive C., Blancard D., Deluc L., Barrieu F., Corio-Costet M.F., Drira N., Hamdi S., Lauvergeat V. Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol. Plant. 2007;131:434–447. doi: 10.1111/j.1399-3054.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 43.Merz P.R., Moser T., Höll J., Kortekamp A., Buchholz G., Zyprian E., Bogs J. The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol. Plant. 2015;153:365–380. doi: 10.1111/ppl.12251. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Z., Shi J., Cao J., He M., Wang Y. VpWRKY3, a biotic and abiotic stress-related transcription factor from the Chinese wild Vitis pseudoreticulata. Plant Cell Rep. 2012;31:2109–2120. doi: 10.1007/s00299-012-1321-1. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Z., Shi J., Xu W., Li H., He M., Xu Y., Xu T., Yang Y., Cao J., Wang Y. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J. Plant Physiol. 2013;170:923–933. doi: 10.1016/j.jplph.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Wu J., Folta K.M., Xie Y., Jiang W., Lu J., Zhang Y. Overexpression of Muscadinia rotundifolia CBF2 gene enhances biotic and abiotic stress tolerance in Arabidopsis. Protoplasma. 2017;254:239–251. doi: 10.1007/s00709-015-0939-6. [DOI] [PubMed] [Google Scholar]
- 47.Yu Y.H., Jiao Z.L., Bian L., Wan Y.T., Yu K.K., Zhang G.H., Guo D.L. Overexpression of Vitis vinifera VvbZIP60 enhances Arabidopsis resistance to powdery mildew via the salicylic acid signaling pathway. Sci. Hortic. 2019;256:108640. doi: 10.1016/j.scienta.2019.108640. [DOI] [Google Scholar]
- 48.Yu Y.H., Bian L., Wan Y.T., Jiao Z.L., Yu K.K., Zhang G.H., Guo D.L. Grape (Vitis vinifera) VvDOF3 functions as a transcription activator and enhances powdery mildew resistance. Plant Physiol. Biochem. 2019;143:183–189. doi: 10.1016/j.plaphy.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Yu Y., Wan Y., Jiao Z., Bian L., Yu K., Zhang G., Guo D. Functional Characterization of Resistance to Powdery Mildew of VvTIFY9 from Vitis vinifera. Int. J. Mol. Sci. 2019;20:4286. doi: 10.3390/ijms20174286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y.H., Li X.Z., Wu Z.J., Chen D.X., Li G.R., Li X.Q., Zhang G.H. VvZFP11, a Cys2His2-type zinc finger transcription factor, is involved in defense responses in Vitis vinifera. Biol. Plant. 2016;60:292–298. doi: 10.1007/s10535-016-0598-2. [DOI] [Google Scholar]
- 51.Coutos-Thévenot P., Poinssot B., Bonomelli A., Yean H., Breda C., Buffard D., Esnault R., Hain R., Boulay M. In vitro tolerance to Botrytis cinerea of grapevine 41B rootstock in transgenic plants expressing the stilbene synthase Vst1 gene under the control of a pathogen-inducible PR 10 promoter. J. Exp. Bot. 2001;52:901–910. doi: 10.1093/jexbot/52.358.901. [DOI] [PubMed] [Google Scholar]
- 52.Dabauza M., Velasco L., Pazos-Navarro M., Pérez-Benito E., Hellín P., Flores P., Gómez-Garay A., Martínez M.C., Lacasa A. Enhanced resistance to Botrytis cinerea in genetically-modified Vitis vinifera L. plants over-expressing the grapevine stilbene synthase gene. Plant Cell Tissue Organ Cult. 2015;120:229–238. doi: 10.1007/s11240-014-0598-x. [DOI] [Google Scholar]
- 53.Fan C., Pu N., Wang X., Wang Y., Fang L., Xu W., Zhang J. Agrobacterium-mediated genetic transformation of grapevine (Vitis vinifera L.) with a novel stilbene synthase gene from Chinese wild Vitis pseudoreticulata. Plant Cell Tissue Organ Cult. 2008;92:197–206. doi: 10.1007/s11240-007-9324-2. [DOI] [Google Scholar]
- 54.Aleynova-Shumakova O.A., Dubrovina A.S., Manyakhin A.Y., Karetin Y.A., Kiselev K.V. VaCPK20 gene overexpression significantly increased resveratrol content and expression of stilbene synthase genes in cell cultures of Vitis amurensis Rupr. Appl. Microbiol. Biotechnol. 2014;98:5541–5549. doi: 10.1007/s00253-014-5625-7. [DOI] [PubMed] [Google Scholar]
- 55.Dai L., Zhou Q., Li R., Du Y., He J., Wang D., Cheng S., Zhang J., Wang Y. Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant Cell Tissue Organ Cult. 2015;121:397–412. doi: 10.1007/s11240-015-0711-9. [DOI] [Google Scholar]
- 56.Jiao Y., Xu W., Duan D., Wang Y., Nick P. A stilbene synthase allele from a Chinese wild grapevine confers resistance to powdery mildew by recruiting salicylic acid signalling for efficient defence. J. Exp. Bot. 2016;67:5841–5856. doi: 10.1093/jxb/erw351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Wang D., Wang F., Huang L., Tian X., van Nocker S., Gao H., Wang X. Expression of the Grape VaSTS19 Gene in Arabidopsis Improves Resistance to Powdery Mildew and Botrytis cinerea but Increases Susceptibility to Pseudomonas syringe pv Tomato DC3000. Int. J. Mol. Sci. 2017;18:2000. doi: 10.3390/ijms18092000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu M., Ma F., Wu F., Jiang C., Wang Y. Expression of stilbene synthase VqSTS6 from wild Chinese Vitis quinquangularis in grapevine enhances resveratrol production and powdery mildew resistance. Planta. 2019;250:1997–2007. doi: 10.1007/s00425-019-03276-2. [DOI] [PubMed] [Google Scholar]
- 59.Zhang G., Yan X., Zhang S., Zhu Y., Zhang X., Qiao H., van Nocker S., Li Z., Wang X. The jasmonate-ZIM domain gene VqJAZ4 from the Chinese wild grape Vitis quinquangularis improves resistance to powdery mildew in Arabidopsis thaliana. Plant Physiol. Biochem. 2019;143:329–339. doi: 10.1016/j.plaphy.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 60.Agüero C.B., Uratsu S.L., Greve C., Powell A.L.T., Labavitch J.M., Meredith C.P., Dandekar A.M. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 2005;6:43–51. doi: 10.1111/j.1364-3703.2004.00262.x. [DOI] [PubMed] [Google Scholar]
- 61.Rubio J., Montes C., Castro Á., Álvarez C., Olmedo B., Muñoz M., Tapia E., Reyes F., Ortega M., Sánchez E., et al. Genetically engineered Thompson Seedless grapevine plants designed for fungal tolerance: Selection and characterization of the best performing individuals in a field trial. Transgenic Res. 2015;24:43–60. doi: 10.1007/s11248-014-9811-2. [DOI] [PubMed] [Google Scholar]
- 62.Yu Y., Xu W., Wang J., Wang L., Yao W., Yang Y., Xu Y., Ma F., Du Y., Wang Y. Erysiphe necator-induced RING finger protein 1 (EIRP1) ahe Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase ctivates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 2013;200:834–846. doi: 10.1111/nph.12418. [DOI] [PubMed] [Google Scholar]
- 63.Wang J., Yao W., Wang L., Ma F., Tong W., Wang C., Bao R., Jiang C., Yang Y., Zhang J., et al. Overexpression of VpEIFP1, a novel F-box/Kelch-repeat protein from wild Chinese Vitis pseudoreticulata, confers higher tolerance to powdery mildew by inducing thioredoxin z proteolysis. Plant Sci. 2017;263:142–155. doi: 10.1016/j.plantsci.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Yu Y., Bian L., Jiao Z., Yu K., Wan Y., Zhang G., Guo D. Molecular cloning and characterization of a grapevine (Vitis vinifera L.) serotonin N-acetyltransferase (VvSNAT2) gene involved in plant defense. BMC Genom. 2019;20:880. doi: 10.1186/s12864-019-6085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y., Li Y., Hou F., Wan D., Cheng Y., Han Y., Gao Y., Liu J., Guo Y., Xiao S., et al. Ectopic expression of Arabidopsis broad-spectrum resistance gene RPW8.2 improves the resistance to powdery mildew in grapevine (Vitis vinifera) Plant Sci. 2018;267:20–31. doi: 10.1016/j.plantsci.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 66.Zhou Q., Dai L., Cheng S., He J., Wang D., Zhang J., Wang Y. A circulatory system useful both for long-term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant Cell Tissue Organ Cult. 2014;118:157–168. doi: 10.1007/s11240-014-0471-y. [DOI] [Google Scholar]
- 67.Jiao L., Zhang Y., Lu J. Overexpression of a stress-responsive U-box protein gene VaPUB affects the accumulation of resistance related proteins in Vitis vinifera ‘Thompson Seedless. Plant Physiol. Biochem. 2017;112:53–63. doi: 10.1016/j.plaphy.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 68.Liu S., Zhang C., Chao N., Lu J., Zhang Y. Cloning, characterization, and functional investigation of VaHAESA from Vitis amurensis inoculated with Plasmopara viticola. Int. J. Mol. Sci. 2018;19:1204. doi: 10.3390/ijms19041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pieterse C.M.J., Zamioudis C., Berendsen R.L., Weller D.M., Van Wees S.C.M., Bakker P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 70.Van Loon L.C., Van Kammen A. Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. “Samsun” and “Samsun NN”. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology. 1970;40:199–211. doi: 10.1016/0042-6822(70)90395-8. [DOI] [PubMed] [Google Scholar]
- 71.Ali S., Mir A., Tyagi A., Bhat J.A., Chandrashekar N., Papolu P.K., Rawat S., Grover A. Identification and comparative analysis of Brassica juncea pathogenesis-related genes in response to hormonal, biotic and abiotic stresses. Acta Physiol. Plant. 2017;39:268. doi: 10.1007/s11738-017-2565-8. [DOI] [Google Scholar]
- 72.Antoniw J.F., Ritter C.E., Pierpoint W.S., Van Loon L.C. Comparison of three pathogenesis-related proteins from plants of two cultivars of tobacco infected with TMV. J. Gen. Virol. 1980;47:79–87. doi: 10.1099/0022-1317-47-1-79. [DOI] [Google Scholar]
- 73.Van Loon L.C. Active Defense Mechanisms in Plants. Springer; Boston, MA, USA: 1982. Regulation of Changes in Proteins and Enzymes Associated with Active Defence against Virus Infection; pp. 247–273. [DOI] [Google Scholar]
- 74.Métraux J.P., Streit L., Staub T. A pathogenesis-related protein in cucumber is a chitinase. Physiol. Mol. Plant Pathol. 1988;33:1–9. doi: 10.1016/0885-5765(88)90038-0. [DOI] [Google Scholar]
- 75.Melchers L.S., Apotheker-de-Groot M., van der Knaap J.A., Ponstein A.S., Sela-Buurlage M.B., Bol J.F., Cornelissen B.J.C., van den Elzen P.J.M., Linthorst H.J.M. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 1994;5:469–480. doi: 10.1046/j.1365-313X.1994.5040469.x. [DOI] [PubMed] [Google Scholar]
- 76.Green T.R., Ryan C.A. Wound-induced proteinase inhibitor in plant leaves: A possible defense mechanism against insects. Science. 1972;175:776–777. doi: 10.1126/science.175.4023.776. [DOI] [PubMed] [Google Scholar]
- 77.Lagrimini L.M., Burkhart W., Moyer M., Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: Molecular analysis and tissue-specific expression. Proc. Natl. Acad. Sci. USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Somssich I.E., Schmelzer E., Bollmann J., Hahlbrock K. Rapid activation by fungal elicitor of genes encoding “pathogenesis-related” proteins in cultured parsley cells. Proc. Natl. Acad. Sci. USA. 1986;83:2427–2430. doi: 10.1073/pnas.83.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terras F.R., Eggermont K., Kovaleva V., Raikhel N.V., Osborn R.W., Kester A., Rees S.B., Torrekens S., Van Leuven F., Vanderleyden J. Small cysteine-rich antifungal proteins from radish: Their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Epple P., Apel K., Bohlmann H. An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 1995;109:813–820. doi: 10.1104/pp.109.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casañal A., Zander U., Muñoz C., Dupeux F., Luque I., Angel Botella M., Schwab W., Valpuesta V., Marquez J.A. The Strawberry Pathogenesis-related 10 (PR-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates. J. Biol. Chem. 2013;288:35322. doi: 10.1074/jbc.M113.501528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L., Wei J., Zou Y., Xu K., Wang Y., Cui L., Xu Y. Molecular Characteristics and Biochemical Functions of VpPR10s from Vitis pseudoreticulata Associated with Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2014;15:19162–19182. doi: 10.3390/ijms151019162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jenssen H., Hamill P., Hancock R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hightower R., Baden C., Penzes E., Dunsmuir P. The expression of cecropin peptide in transgenic tobacco does not confer resistance to Pseudomonas syringae pv tabaci. Plant Cell Rep. 1994;13:295–299. doi: 10.1007/BF00233324. [DOI] [PubMed] [Google Scholar]
- 85.Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10:71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 86.Singh K.B., Foley R.C., Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 87.Marchive C., Léon C., Kappel C., Coutos-Thévenot P., Corio-Costet M.F., Delrot S., Lauvergeat V. Over-Expression of VvWRKY1 in Grapevines Induces Expression of Jasmonic Acid Pathway-Related Genes and Confers Higher Tolerance to the Downy Mildew. PLoS ONE. 2013;8:e54185. doi: 10.1371/journal.pone.0054185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 89.Costenaro da Silva D., da Silveira Falavigna V., Fasoli M., Buffon V., Denardi Porto D., Joannis Pappas Jr G., Pezzotti M., Pasquali G., Fernando Revers L. Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development. Hortic. Res. 2016;3 doi: 10.1038/hortres.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hasan M.M., Bae H. An overview of stress-induced resveratrol synthesis in grapes: Perspectives for resveratrol-enriched grape products. Molecules. 2017;22:294. doi: 10.3390/molecules22020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adrian M., Jeandet P. Effects of resveratrol on the ultrastructure of Botrytis cinerea conidia and biological significance in plant/pathogen interactions. Fitoterapia. 2012;83:1345–1350. doi: 10.1016/j.fitote.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 92.Gray D.J., Li Z.T., Dhekney S.A. Precision breeding of grapevine (Vitis vinifera L.) for improved traits. Plant Sci. 2014;228:3–10. doi: 10.1016/j.plantsci.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 93.Arnao M.B., Hernández-Ruiz J. Melatonin: Plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 2014;19:789–797. doi: 10.1016/j.tplants.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 94.Xiao S., Ellwood S., Calis O., Patrick E., Li T., Coleman M., Turner J.G. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- 95.Bhat A., Ryu C.-M. Plant Perceptions of Extracellular DNA and RNA. Mol. Plant. 2016;9:956–958. doi: 10.1016/j.molp.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 96.Huang C.Y., Wang H., Hu P., Hamby R., Jin H. Small RNAs—Big Players in Plant-Microbe Interactions. Cell Host Microbe. 2019;26:173–182. doi: 10.1016/j.chom.2019.07.021. [DOI] [PubMed] [Google Scholar]
- 97.Hou Y., Zhai Y., Feng L., Karimi H.Z., Rutter B.D., Zeng L., Choi D.S., Zhang B., Gu W., Chen X., et al. A Phytophthora Effector Suppresses Trans-Kingdom RNAi to Promote Disease Susceptibility. Cell Host Microbe. 2019;25:153–165. doi: 10.1016/j.chom.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dalakouras A., Wassenegger M., Dadami E., Ganopoulos I., Pappas M.L., Papadopoulou K. Update on Exogenous Application of RNAi Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants 1[OPEN] Plant Physiol. Ò. 2020;182:38–50. doi: 10.1104/pp.19.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cai Q., Qiao L., Wang M., He B., Lin F.-M., Palmquist J., Huang S.-D., Jin H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360:1126–1129. doi: 10.1126/science.aar4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang M., Weiberg A., Lin F.M., Thomma B.P.H.J., Da Huang H., Jin H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants. 2016;2:16151. doi: 10.1038/nplants.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nowara D., Schweizer P., Gay A., Lacomme C., Shaw J., Ridout C., Douchkov D., Hensel G., Kumlehn J. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22:3130–3141. doi: 10.1105/tpc.110.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pliego C., Nowara D., Bonciani G., Gheorghe D.M., Xu R., Surana P., Whigham E., Nettleton D., Bogdanove A.J., Wise R.P., et al. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant-Microbe Interact. 2013;26:633–642. doi: 10.1094/MPMI-01-13-0005-R. [DOI] [PubMed] [Google Scholar]
- 103.Whigham E., Qi S., Mistry D., Surana P., Xu R., Fuerst G., Pliego C., Bindschedler L.V., Spanu P.D., Dickerson J.A., et al. Broadly conserved fungal effector BEC1019 suppresses host cell death and enhances pathogen virulence in powdery mildew of barley (Hordeum vulgare L.) Mol. Plant-Microbe Interact. 2015;28:968–983. doi: 10.1094/MPMI-02-15-0027-FI. [DOI] [PubMed] [Google Scholar]
- 104.Koch A., Kumar N., Weber L., Keller H., Imani J., Kogel K.H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA. 2013;110:19324–19329. doi: 10.1073/pnas.1306373110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ghag S.B., Shekhawat U.K.S., Ganapathi T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014;12:541–553. doi: 10.1111/pbi.12158. [DOI] [PubMed] [Google Scholar]
- 106.Cheng W., Song X.-S., Li H.-P., Cao L.-H., Sun K., Qiu X.-L., Xu Y.-B., Yang P., Huang T., Zhang J.-B., et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015;13:1335–1345. doi: 10.1111/pbi.12352. [DOI] [PubMed] [Google Scholar]
- 107.Panwar V., McCallum B., Bakkeren G. Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 2013;81:595–608. doi: 10.1007/s11103-013-0022-7. [DOI] [PubMed] [Google Scholar]
- 108.Zhang T., Jin Y., Zhao J.-H., Gao F., Zhou B.-J., Fang Y.-Y., Guo H.-S. Host-Induced Gene Silencing of the Target Gene in Fungal Cells Confers Effective Resistance to the Cotton Wilt Disease Pathogen Verticillium dahliae. Mol. Plant. 2016;9:939–942. doi: 10.1016/j.molp.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 109.Vega-Arreguín J.C., Jalloh A., Bos J.I., Moffett P. Recognition of an Avr3a homologue plays a major role in mediating nonhost resistance to phytophthora capsici in Nicotiana species. Mol. Plant-Microbe Interact. 2014;27:770–780. doi: 10.1094/MPMI-01-14-0014-R. [DOI] [PubMed] [Google Scholar]
- 110.Sanju S., Siddappa S., Thakur A., Shukla P.K., Srivastava N., Pattanayak D., Sharma S., Singh B.P. Host-mediated gene silencing of a single effector gene from the potato pathogen Phytophthora infestans imparts partial resistance to late blight disease. Funct. Integr. Genom. 2015;15:697–706. doi: 10.1007/s10142-015-0446-z. [DOI] [PubMed] [Google Scholar]
- 111.Jahan S.N., Åsman A.K.M., Corcoran P., Fogelqvist J., Vetukuri R.R., Dixelius C. Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J. Exp. Bot. 2015;66:2785–2794. doi: 10.1093/jxb/erv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Govindarajulu M., Epstein L., Wroblewski T., Michelmore R.W. Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 2015;13:875–883. doi: 10.1111/pbi.12307. [DOI] [PubMed] [Google Scholar]
- 113.Panstruga R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 2005;33:389–392. doi: 10.1042/BST0330389. [DOI] [PubMed] [Google Scholar]
- 114.Pessina S., Lenzi L., Perazzolli M., Campa M., Dalla Costa L., Urso S., Valè G., Salamini F., Velasco R., Malnoy M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 2016;3:16016. doi: 10.1038/hortres.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sabbadini S., Capriotti L., Molesini B., Pandolfini T., Navacchi O., Limera C., Ricci A., Mezzetti B. Comparison of regeneration capacity and Agrobacterium-mediated cell transformation efficiency of different cultivars and rootstocks of Vitis spp. via organogenesis. Sci. Rep. 2019;9:582. doi: 10.1038/s41598-018-37335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dubrovina A., Aleynova O., Kalachev A., Suprun A., Ogneva Z., Kiselev K. Induction of Transgene Suppression in Plants via External Application of Synthetic dsRNA. Int. J. Mol. Sci. 2019;20:1585. doi: 10.3390/ijms20071585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Koch A., Biedenkopf D., Furch A., Weber L., Rossbach O., Abdellatef E., Linicus L., Johannsmeier J., Goesmann J.L., Cardoza V., et al. An RNAi-based Control of Fusarium graminearum Infections Through Spraying of Long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016;12:e1005901. doi: 10.1371/journal.ppat.1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McLoughlin A.G., Wytinck N., Walker P.L., Girard I.J., Rashid K.Y., De Kievit T., Fernando W.G.D., Whyard S., Belmonte M.F. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-25434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gu K.X., Song X.S., Xiao X.M., Duan X.X., Wang J.X., Duan Y.B., Hou Y.P., Zhou M.G. A β 2 -tubulin dsRNA derived from Fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance. Pestic. Biochem. Physiol. 2019;153:36–46. doi: 10.1016/j.pestbp.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 120.Cagliari D., Avila dos Santos E., Dias N., Smagghe G., Zotti M. Modulating Gene Expression—Abridging the RNAi and CRISPR-Cas9 Technologies [Working Title] IntechOpen; London, UK: 2019. Nontransformative Strategies for RNAi in Crop Protection. [DOI] [Google Scholar]
- 121.Sang H., Kim J. Il Advanced strategies to control plant pathogenic fungi by host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS) Plant Biotechnol. Rep. 2020;14:1–8. doi: 10.1007/s11816-019-00588-3. [DOI] [Google Scholar]
- 122.Dalakouras A., Jarausch W., Buchholz G., Bassler A., Braun M., Manthey T., Krczal G., Wassenegger M. Delivery of Hairpin RNAs and Small RNAs Into Woody and Herbaceous Plants by Trunk Injection and Petiole Absorption. Front. Plant Sci. 2018;9:1253. doi: 10.3389/fpls.2018.01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dubrovina A.S., Kiselev K.V. Exogenous RNAs for Gene Regulation and Plant Resistance. Int. J. Mol. Sci. 2019;20:2282. doi: 10.3390/ijms20092282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Das P.R., Sherif S.M. Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020;11:946. doi: 10.3389/fpls.2020.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang M., Dean R.A. Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 2020;21:589–601. doi: 10.1111/mpp.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Song X.S., Gu K.X., Duan X.X., Xiao X.M., Hou Y.P., Duan Y.B., Wang J.X., Zhou M.G. A myosin5 dsRNA that reduces the fungicide resistance and pathogenicity of Fusarium asiaticum. Pestic. Biochem. Physiol. 2018;150:1–9. doi: 10.1016/j.pestbp.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 127.Dalakouras A., Wassenegger M., McMillan J.N., Cardoza V., Maegele I., Dadami E., Runne M., Krczal G., Wassenegger M. Induction of silencing in plants by high-pressure spraying of In vitro-synthesized small RNAs. Front. Plant Sci. 2016;7:1327. doi: 10.3389/fpls.2016.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nerva L., Sandrini M., Gambino G., Chitarra W. Double-Stranded RNAs (dsRNAs) as a Sustainable Tool against Gray Mold (Botrytis cinerea) in Grapevine: Effectiveness of Different Application Methods in an Open-Air Environment. Biomolecules. 2020;10:200. doi: 10.3390/biom10020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bennett M., Deikman J., Hendrix B., Iandolino A. Barriers to Efficient Foliar Uptake of dsRNA and Molecular Barriers to dsRNA Activity in Plant Cells. Front. Plant Sci. 2020;11:816. doi: 10.3389/fpls.2020.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Demirci Y., Zhang B., Unver T. CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant genome editing. J. Cell. Physiol. 2018;233:1844–1859. doi: 10.1002/jcp.25970. [DOI] [PubMed] [Google Scholar]
- 131.Wang X., Tu M., Wang D., Liu J., Li Y., Li Z., Wang Y., Wang X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018;16:844–855. doi: 10.1111/pbi.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ren C., Liu X., Zhang Z., Wang Y., Duan W., Li S., Liang Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.) Sci. Rep. 2016;6:32289. doi: 10.1038/srep32289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 134.Pan C., Ye L., Qin L., Liu X., He Y., Wang J., Chen L., Lu G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016;6:24765. doi: 10.1038/srep24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakajima I., Azuma A., Onoue N., Moriguchi T., Yamamoto T., Toki S., Endo M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE. 2017;12:e0177966. doi: 10.1371/journal.pone.0177966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ren F., Ren C., Zhang Z., Duan W., Lecourieux D., Li S., Liang Z. Efficiency Optimization of CRISPR/Cas9-Mediated Targeted Mutagenesis in Grape. Front. Plant Sci. 2019;10:612. doi: 10.3389/fpls.2019.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ma X., Zhu Q., Chen Y., Liu Y.G. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant. 2016;9:961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 138.Osakabe Y., Liang Z., Ren C., Nishitani C., Osakabe K., Wada M., Komori S., Malnoy M., Velasco R., Poli M., et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018;13:2844–2863. doi: 10.1038/s41596-018-0067-9. [DOI] [PubMed] [Google Scholar]
- 139.Bertini E., Tornielli G.B., Pezzotti M., Zenoni S. Regeneration of plants from embryogenic callus-derived protoplasts of Garganega and Sangiovese grapevine (Vitis vinifera L.) cultivars. Plant Cell Tissue Organ Cult. 2019;138:239–246. doi: 10.1007/s11240-019-01619-1. [DOI] [Google Scholar]
- 140.Malnoy M., Viola R., Jung M.-H., Koo O.-J., Kim S., Kim J.-S., Velasco R., Nagamangala Kanchiswamy C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Giacomelli L., Zeilmaker T., Dalla Costa L., Malnoy M., Rouppe van der Voort J., Moser C. LXII SIGA Annual Congress: Plant Development and Crop Productivity for Sustainable Agriculture. SIGA; Verona, Italy: 2018. Generation of mildew-resistant grapevine clones via genome editing: Potentials and hurdles; p. 904570. [DOI] [Google Scholar]
- 142.Zimny T., Sowa S., Tyczewska A., Twardowski T. Certain new plant breeding techniques and their marketability in the context of EU GMO legislation—Recent developments. N Biotechnol. 2019;51:49–56. doi: 10.1016/j.nbt.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 143.Dalakouras A., Wassenegger M., Dadami E., Ganopoulos I., Pappas M., Papadopoulou K.K. GMO-free RNAi: Exogenous application of RNA molecules in plants. Plant Physiol. 2019:00570. doi: 10.1104/pp.19.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yang S., Fresnedo-Ramírez J., Wang M., Cote L., Schweitzer P., Barba P., Takacs E.M., Clark M., Luby J., Manns D.C., et al. A next-generation marker genotyping platform (AmpSeq) in heterozygous crops: A case study for marker-assisted selection in grapevine. Hortic. Res. 2016;3:16002. doi: 10.1038/hortres.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Myles S. Improving fruit and wine: What does genomics have to offer? Trends Genet. 2013;29:190–196. doi: 10.1016/j.tig.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 146.Riaz S., Tenscher A.C., Graziani R., Krivanck A.F., Ramming D.W., Andrew Walkcrl M., Kahica K., Dervishian G., Ng D. Using Marker-Assisted Selection to Breed Pierce’s Disease-Resistant Grapes. Am. J. Enol. Vitic. 2009;60:199–207. [Google Scholar]
- 147.Hartung F., Schiemann J. Precise plant breeding using new genome editing techniques: Opportunities, safety and regulation in the EU. Plant J. 2014;78:742–752. doi: 10.1111/tpj.12413. [DOI] [PubMed] [Google Scholar]
- 148.Schouten H.J., Krens F.A., Jacobsen E. Cisgenic plants are similar to traditionally bred plants. EMBO Rep. 2006;7:750–753. doi: 10.1038/sj.embor.7400769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rommens C.M., Haring M.A., Swords K., Davies H.V., Belknap W.R. The intragenic approach as a new extension to traditional plant breeding. Trends Plant Sci. 2007;12:397–403. doi: 10.1016/j.tplants.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 150.Joshi S.G., Schaart J.G., Groenwold R., Jacobsen E., Schouten H.J., Krens F.A. Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol. Biol. 2011;75:579–591. doi: 10.1007/s11103-011-9749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zotti M., dos Santos E.A., Cagliari D., Christiaens O., Taning C.N.T., Smagghe G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018;74:1239–1250. doi: 10.1002/ps.4813. [DOI] [PubMed] [Google Scholar]
- 152.Zischewski J., Fischer R., Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017;35:95–104. doi: 10.1016/j.biotechadv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 153.Lusk J.L., Rozan A. Consumer acceptance of ingenic foods. Biotechnol. J. 2006;1:1433–1434. doi: 10.1002/biot.200600187. [DOI] [PubMed] [Google Scholar]
- 154.Frewer L.J. Consumer acceptance and rejection of emerging agrifood technologies and their applications. Eur. Rev. Agric. Econ. 2017;44:683–704. doi: 10.1093/erae/jbx007. [DOI] [Google Scholar]
- 155.Delwaide A.C., Nalley L.L., Dixon B.L., Danforth D.M., Nayga R.M., Van Loo E.J., Verbeke W. Revisiting GMOs: Are there differences in European consumers’ acceptance and valuation for cisgenically vs transgenically bred rice? PLoS ONE. 2015;10:e0126060. doi: 10.1371/journal.pone.0126060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lucht J. Public Acceptance of Plant Biotechnology and GM Crops. Viruses. 2015;7:4254–4281. doi: 10.3390/v7082819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Malyska A., Bolla R., Twardowski T. The Role of Public Opinion in Shaping Trajectories of Agricultural Biotechnology. Trends Biotechnol. 2016;34:530–534. doi: 10.1016/j.tibtech.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 158.Telem R., Wani S., Singh N., Nandini R., Sadhukhan R., Bhattacharya S., Mandal N. Cisgenics—A Sustainable Approach for Crop Improvement. Curr. Genom. 2013;14:468–476. doi: 10.2174/13892029113146660013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jacobsen E., Schouten H.J. Cisgenesis, a New Tool for Traditional Plant Breeding, Should be Exempted from the Regulation on Genetically Modified Organisms in a Step by Step Approach. Potato Res. 2008;51:75–88. doi: 10.1007/s11540-008-9097-y. [DOI] [Google Scholar]
- 160.Arpaia S., Christiaens O., Giddings K., Jones H., Mezzetti B., Moronta-Barrios F., Perry J.N., Sweet J.B., Taning C.N.T., Smagghe G., et al. Biosafety of GM Crop Plants Expressing dsRNA: Data Requirements and EU Regulatory Considerations. Front. Plant Sci. 2020;11:940. doi: 10.3389/fpls.2020.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Casacuberta J.M., Devos Y., du Jardin P., Ramon M., Vaucheret H., Nogué F. Biotechnological uses of RNAi in plants: Risk assessment considerations. Trends Biotechnol. 2015;33:145–147. doi: 10.1016/j.tibtech.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 162.Mezzetti B., Smagghe G., Arpaia S., Christiaens O., Dietz-Pfeilstetter A., Jones H., Kostov K., Sabbadini S., Opsahl-Sorteberg H.-G., Ventura V., et al. RNAi: What is its position in agriculture? J. Pest Sci. 2020;1:3. doi: 10.1007/s10340-020-01238-2. [DOI] [Google Scholar]
- 163.Papadopoulou N., Devos Y., Álvarez-Alfageme F., Lanzoni A., Waigmann E. Risk Assessment Considerations for Genetically Modified RNAi Plants: EFSA’s Activities and Perspective. Front. Plant Sci. 2020;11:445. doi: 10.3389/fpls.2020.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shew A.M., Danforth D.M., Nalley L.L., Nayga R.M., Tsiboe F., Dixon B.L. New innovations in agricultural biotech: Consumer acceptance of topical RNAi in rice production. Food Control. 2017;81:189–195. doi: 10.1016/j.foodcont.2017.05.047. [DOI] [Google Scholar]
- 165.El-Mounadi K., Morales-Floriano M.L., Garcia-Ruiz H. Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant Sci. 2020;11:56. doi: 10.3389/fpls.2020.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ishii T., Araki M. Consumer acceptance of food crops developed by genome editing. Plant Cell Rep. 2016;35:1507–1518. doi: 10.1007/s00299-016-1974-2. [DOI] [PubMed] [Google Scholar]
- 167.Voytas D.F., Gao C. Precision Genome Engineering and Agriculture: Opportunities and Regulatory Challenges. PLoS Biol. 2014;12:e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]