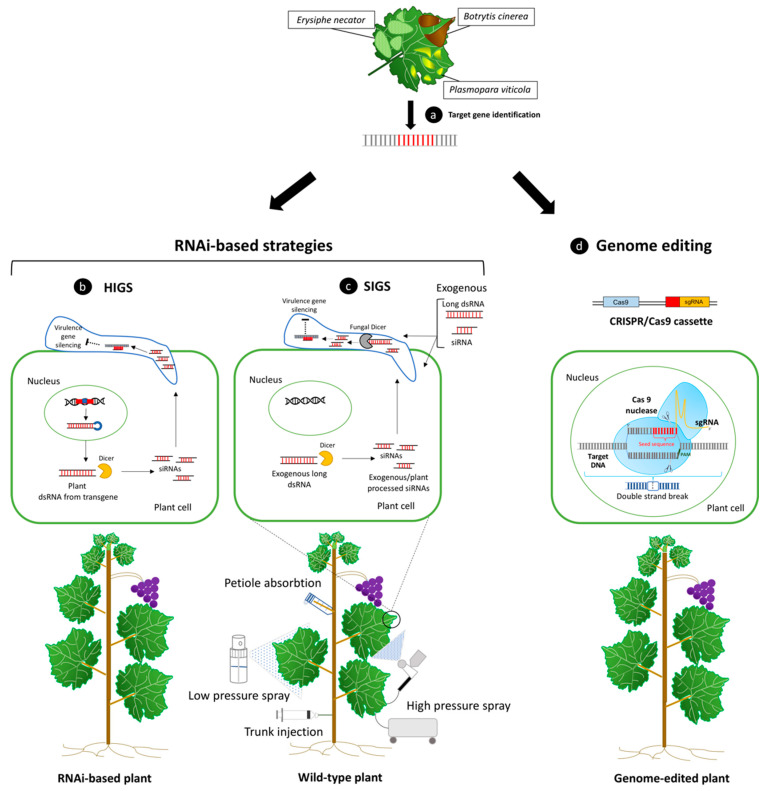
Figure 2.
In addition to trans/cisgenesis methods, the expression of RNAi gene constructs in the plant, the exogenous applications of double strand RNA (dsRNA) molecules targeting host/pathogen genes, or plant genome editing, represent valid alternatives to enhance plant immunity during pathogenesis. (a) Candidate genes capable of limiting pathogen aggression or improve plant defense responses can be identified during the infection processes caused by the fungal and oomycetes causal agents of the most impactful diseases for grapevine production. RNAi-based strategies can be exploited to improve plant defense by providing dsRNAs to the plant cell through the expression of an introgressed hairpin-based gene construct in the plant genome, or through their delivery by exogenous application. (b) In host-induced gene silencing (HIGS), as a result of transcription of an RNAi sequence, a long dsRNA molecule is formed. When this molecule is recognized by Dicer-like protein, it is cleaved into siRNAs, which can knockdown related target gene expression [100]. (c) A transgenic-free procedure in which dsRNAs are directly sprayed on the surface of plants and pathogens is known as spray-induced gene silencing (SIGS). These molecules can be absorbed by both types of cells, and, depending on the delivery method used, dsRNAs can be processed by either the fungal/oomycetes and host RNAi machinery, leading to virulence gene knockdown and reduction in pathogen detrimental effects. Low-pressure spray, high-pressure spray, petiole adsorption, and trunk injection of dsRNAs represent some of the different available exogenous dsRNA delivery methods to confer plant protection against different plant pathogens, included fungi [143]. d) CRISPR/Cas9 system can be used for inducing targeted genome editing in plants, including the inactivation of specific plant susceptibility genes expression, which can help to regulate plant–pathogen interaction processes and disease resistance enhancement. Cas 9 protein complex is guided by artificially designed single guide RNA molecule (sgRNA) and leads to double-strand breaks (DSBs) of targeted DNA. SgRNA contains a seed sequence (around 8–12 bp, shown in red) complementary to target DNA that guides the binding of the Cas 9 protein to the target genomic sequence. The site of cleavage takes place three nucleotides upstream to the protospacer adjacent motif (PAM, shown in green) [20].