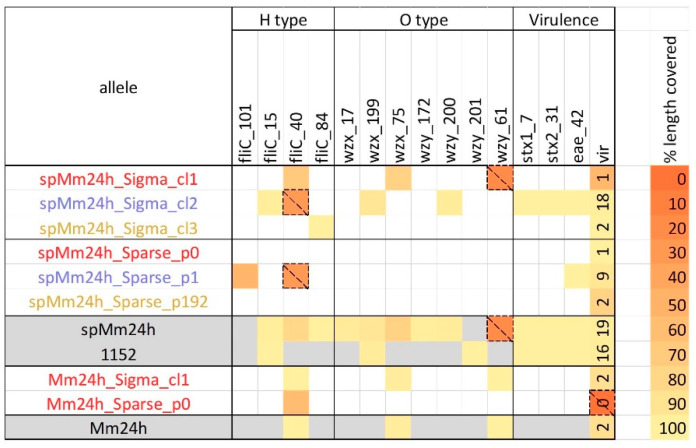
Figure 2.
Gene detection of the O- and H-type serotyping and virulence genes performed on the clusters detected by Sigma and Sparse in the spiked (spMm24h) and unspiked (Mm24h) enriched minced meat samples. The table includes only the three largest clusters from spMm24h and only the first largest cluster from Mm24h, as none of the smaller clusters generated by any the two tools contained any of the monitored genes. In addition to the clusters generated by Sigma and Sparse, for comparison reasons, gene detection was performed on the reads obtained for the whole metagenomic samples, Mm24h and spMm24h, and on those of isolate TIAC1152 that was used for spiking. The Shiga toxin-producing Escherichia coli (STEC)-specific virulence genes (stx and eae), are displayed separately, while for the remaining virulence genes (vir), only the total number of the detected genes is shown (see Figure S3 for more detailed information). Cell color represents the percentage of the allele length covered by reads (%). Only alleles covered for more than 50% at least once are included in the table. Thereby, alleles that are covered below 50% are encased with dashed lines, and are not considered during interpretation of the results.