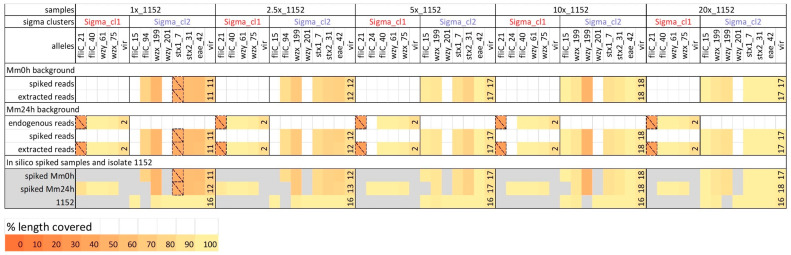
Figure 4.
Strain-level analysis of in silico spiked metagenomic samples containing the strain TIAC1152 at different coverages: gene detection. Isolate TIAC1152 reads were down-sampled to different coverages (spiked reads) and spiked into the following metagenomic backgrounds: the non-enriched minced meat sample containing no endogenous E. coli strains (Mm0h background) and the enriched minced meat sample containing one more prevalent (endogenous reads) and one negligible endogenous E. coli strain according to Sigma (the latter strain contained no virulence or serotyping genes and is therefore omitted) (Mm24h background). The reads attributed to different Sigma clusters and thus presumably belonging to the different strains were extracted from the resulting in silico spiked metagenomic samples (extracted reads), and gene detection of the O- and H-type serotyping genes, STEC-specific virulence genes (stx and eae) and remaining virulence genes (vir) was performed. For the latter, only the total number of detected genes is shown (see Figure S4 for more details). The detected genes are grouped according to the Sigma clusters, in which the corresponding reads were retrieved. The line “endogenous reads” in the Mm24h background shows the genes observed in the main cluster extracted by Sigma from the unspiked Mm24h sample (Mm24h_Sigma_cl1). The lowest section of the table shows genes observed in the whole in silico spiked metagenomic samples prior to Sigma analysis (spiked Mm0h and spiked Mm24h) and in the non-downsampled sequencing data of isolate TIAC1152, the latter showing which serotyping and virulence gene alleles are expected for isolate TIAC1152. Cell color represents the percentage of the allele length covered by reads (%). Only alleles covered for more than 50% at least once are included in the table. Thereby, alleles that are covered below 50% are encased with dashed lines, and are not considered during interpretation of the results.