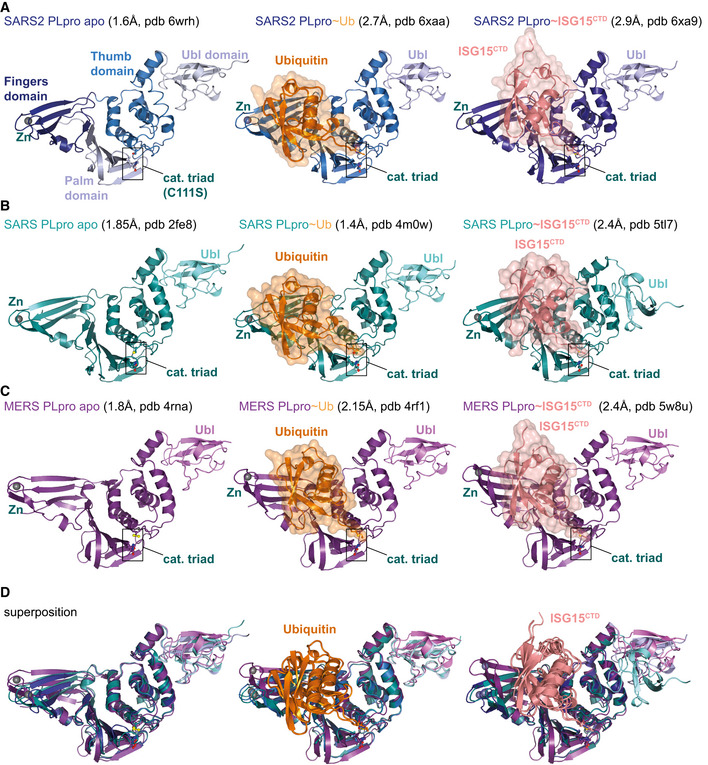
Figure EV2. Selection of available structures of coronavirus PLpro.
-
Aleft, the unpublished structure of apo SARS2 PLpro (pdb 6wrh, 1.6 Å, determined by the Centre for Structural Genomics of Infectious Disease (CSGID)) is coloured in analogy with Figs 1E and EV1, indicating Thumb, Fingers and Palm subdomain. The PLpro fold forms an open right hand that holds the ubiquitin fold, guiding its C‐terminus into the active site. PLpro contains an N‐terminal Ubl domain of unknown function, and this domain can vary in orientation. The high‐resolution structure was generated with a catalytic Cys to Ser mutation; a more common catalytic Cys to Ala mutant destabilises PLpro (Fig EV3D). Middle, structure of SARS2 PLpro bound to ubiquitin (orange, covered by a semi‐transparent surface). Right, structure of SARS2 PLpro bound to the C‐terminal domain of ISG15 (ISG15CTD, salmon, under a semi‐transparent surface). Also see Fig 1. In ubiquitin and ISG15CTD complexes, propargylamide‐based suicide probes (Ekkebus et al, 2013) covalently modify catalytic Cys111.
- B
- C
-
DSuperpositions of PLpro structures with S1 site occupied by different modifiers. Overall, PLpro shows high similarity, extending to the position and orientation of the N‐terminal Ubl domain, with the notable exception of a distinct position of the Ubl in the SARS complex with ISG15CTD (pdb 5tl7 (Daczkowski et al, 2017a)). The second most variable region concerns the Fingers subdomain, which shows varying degrees of “openness.” Superposition shows that the structures of SARS and SARS2 bound to individual modifiers are highly similar, and the modifiers adapt identical orientations and engage in similar interactions with PLpro. MERS PLpro seems to vary on the theme of ubiquitin versus ISG15 recognition, by binding both modifiers similarly. In the MERS ubiquitin complex, the Fingers are more closed, and the ubiquitin is pushed towards the Thumb domain, to adopt a similar orientation and interaction as seen for ISG15 bound to MERS. MERS PLpro ubiquitin complexes have been determined with “open” and more “closed” Fingers (Bailey‐Elkin et al, 2014).
