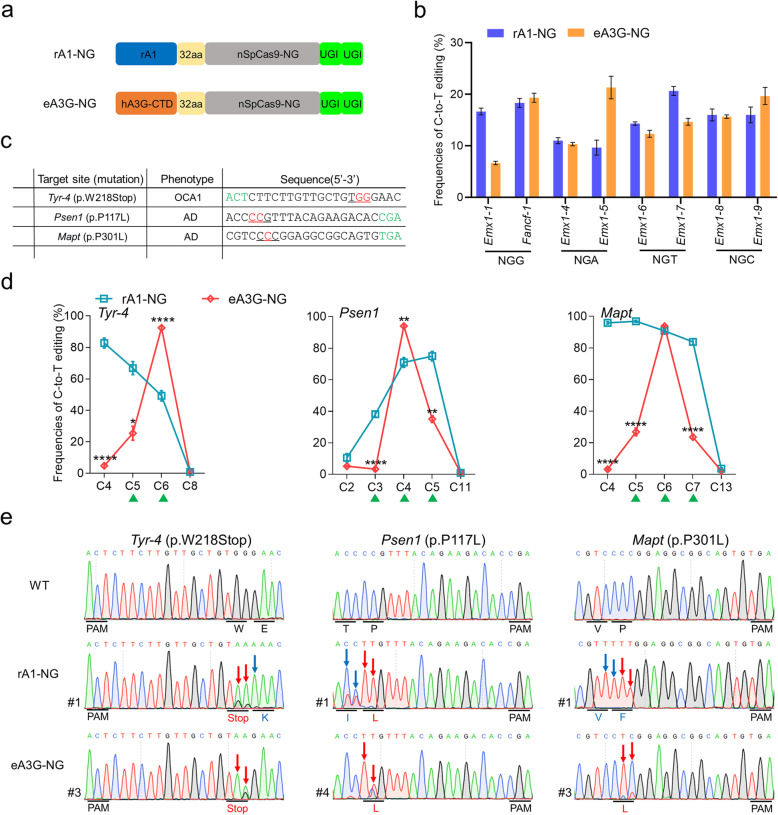
Fig. 5.
Expanded targeting scope using optimized eA3G-NG fusion in rabbit embryos. a Schematic representation of rA1-NG and eA3G-NG architecture. b Comparison of the base editing frequencies between rA1-NG and eA3G-NG with NGN PAMs in HEK293T cells. Values and error bars reflect the mean ± s.e.m. of three independent biological replicates. c The three target-site sequences within NG PAMs. Target sequence (black), PAM region (green), target base (red), and mutant amino acid (underlined). d Comparison of the editing frequencies of single C-to-T conversions between rA1-NG and eA3G-NG at three sites with NG PAMs in rabbit embryos. CCC context (green triangle). n = ~ 6 blastocysts. e Representative sequencing chromatograms of edited rabbit blastocyst at three target sites using rA1-NG and eA3G-NG systems. Targeted mutations (red arrows) and bystander mutations (blue arrows). The relevant codon identities at the target site are presented under the DNA sequence