Figure 1.
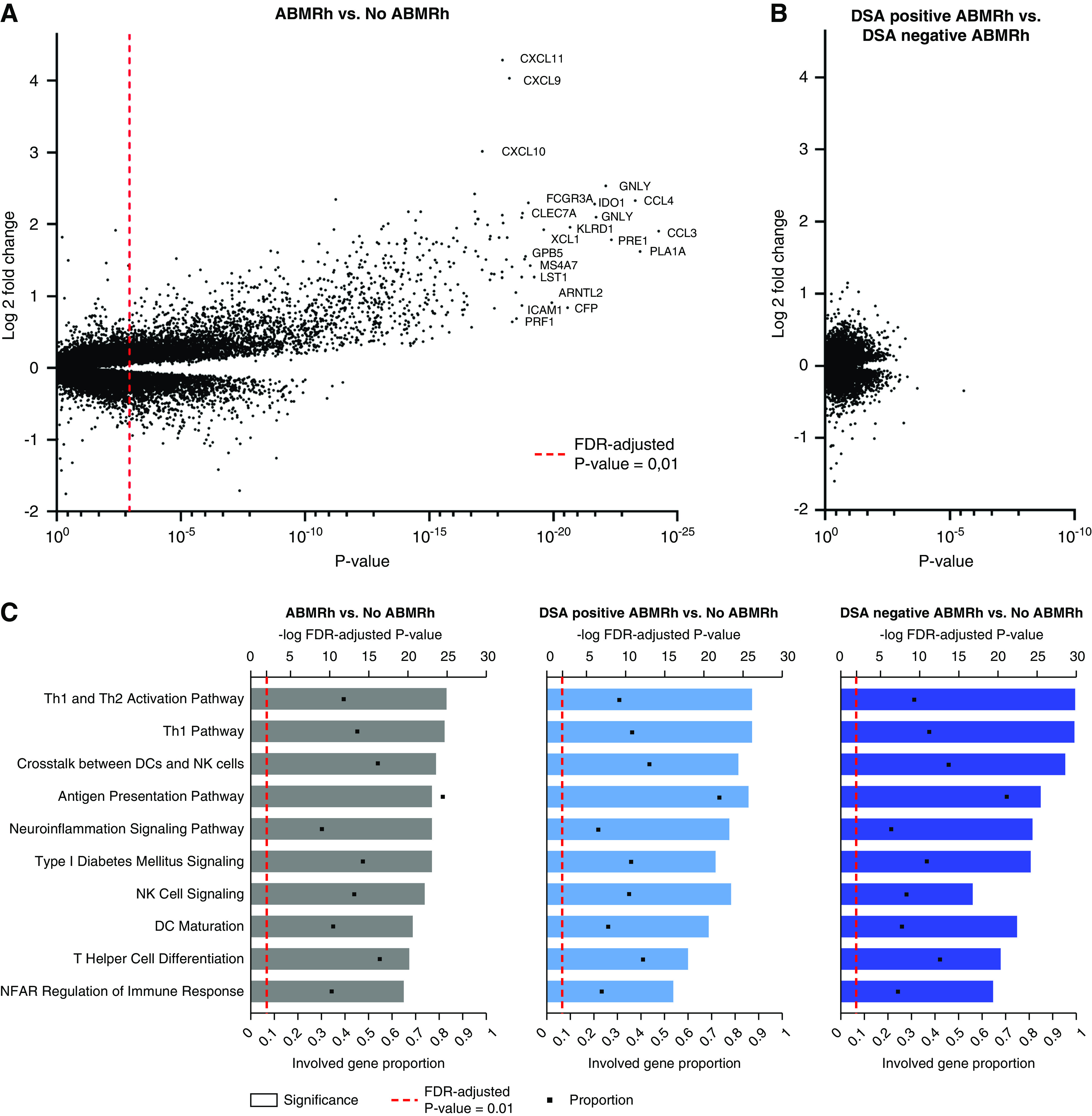
HLA-DSA-negative ABMRh exhibits similar transcriptional changes as HLA-DSA-positive ABMRh. (A) Differential gene expression analysis between ABMRh (n=56) and no ABMRh (n=168). Genes can be represented by multiple probe sets within a single microarray. FDR-adjusted P value <0.01 was considered significant (dashed red line). (B) Differential gene expression analysis between HLA-DSA–negative ABMRh (n=26) and HLA-DSA–positive ABMRh (n=30). No probe sets were found to differ between these two groups after FDR adjustment, despite a clear distinction between ABMRh cases and other biopsy specimens as presented in (A). (C) Ingenuity pathway analysis of differentially expressed genes in ABMRh, HLA-DSA–positive ABMRh, or HLA-DSA–negative ABMRh versus other biopsy specimens. The top ten upregulated pathways in ABMRh were preserved in both ABMRh subtypes. Gene proportion denotes the fraction of known genes within the analyzed pathway that are significantly overexpressed. DCs, dendritic cells; NFAT, nuclear factor of activated T cells; Th1, T helper type 1 cell.
