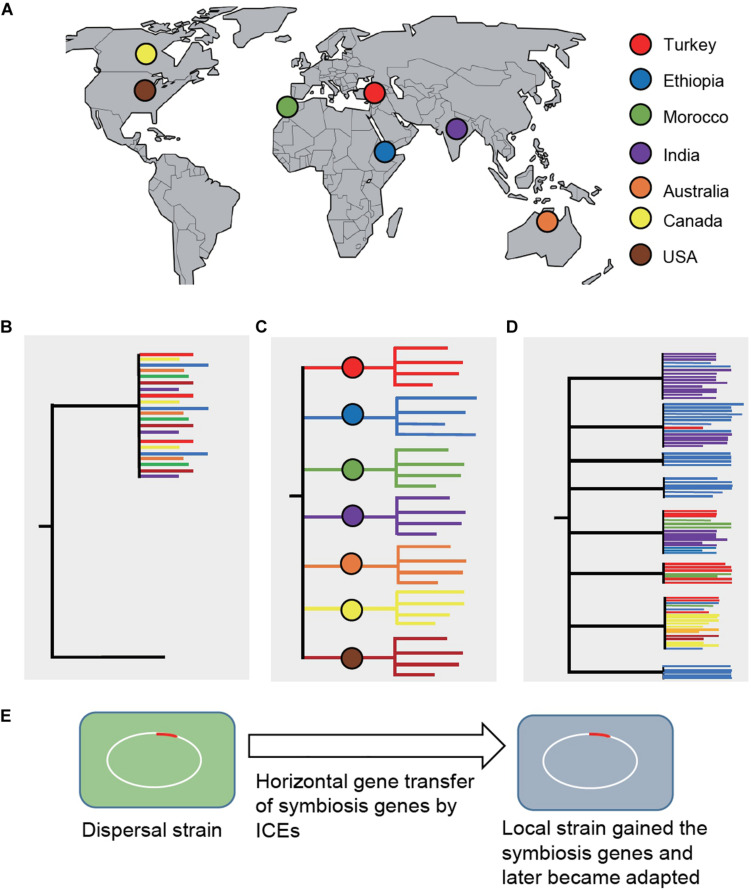
FIGURE 1.
Phylogenetic analysis of Mesorhizobium strains sampled across the chickpea’s native and introduced ranges (Greenlon et al., 2019). (A) Geographic locations (countries) where the strains were sampled, including the chickpea’s domestication center, Turkey. (B) A phylogenetic diagram showing clustering of bacterial strains from the native and introduced ranges under the dispersal (co-introduction) model. (C) A phylogenetic diagram showing that bacterial strains would cluster based on their geographic locations and show a biogeographical pattern if the domesticated plants acquired novel symbionts in their introduced locations. (D) If both co-introduction and co-adaptation happened, the tree would show mixed patterns of (A) and (B), which is the case revealed by the study of chickpea’s symbionts (Greenlon et al., 2019). Trees are not drawn in scale. (E) Strains in the introduced ranges might have acquired symbiosis genes from dispersed strains and subsequently became adapted and outcompeted the dispersal strains. Chromosomal symbiosis genes in Mesorhizobium are encoded on mobile integrative and conjugative elements (ICEs). ICEs can excise from the chromosome that can transfer and integrate into the chromosome of a recipient strain.