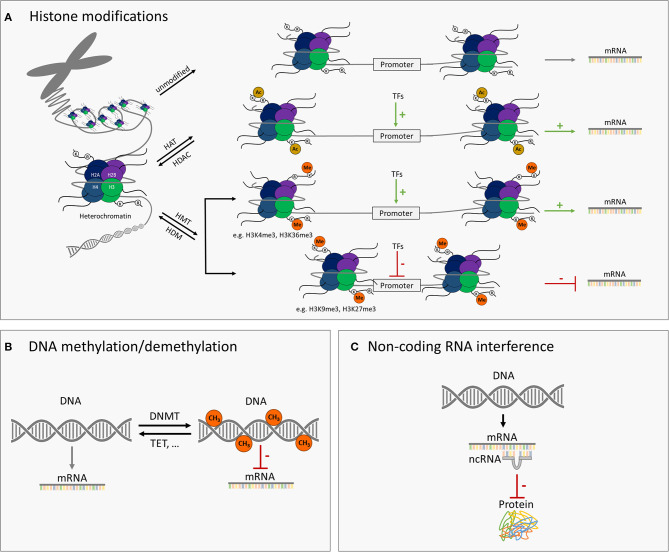
Figure 1.
Schematic illustration of major epigenetic modifications. (A) Modification of histones such as histone acetylation/deacetylation via histone acetyltransferases (HATs)/histone deacetylases (HDAC) and methylation/demethylation via histone methyltransferases (HMT)/histone demethylases (HDM) can either activate or repress the target gene transcription. Histone acetylation is typically associated with higher expression of the gene. Histone methylation can be related to either higher or lower transcriptional activity, depending on the amino acid residue modified and the number of methyl groups added. (B) DNA methylation or demethylation of genomic DNA through DNA methyltransferases (DNMT) or ten-eleven translocation (TET) enzymes and others, respectively. Higher level of DNA methylation is typically associated with lower transcriptional activity of the respective gene. (C) MicroRNAs (miRNAs) and further small non-coding RNAs can interfere with gene expression through base pairing with messenger RNAs and thus inhibiting their translation into the encoded protein.