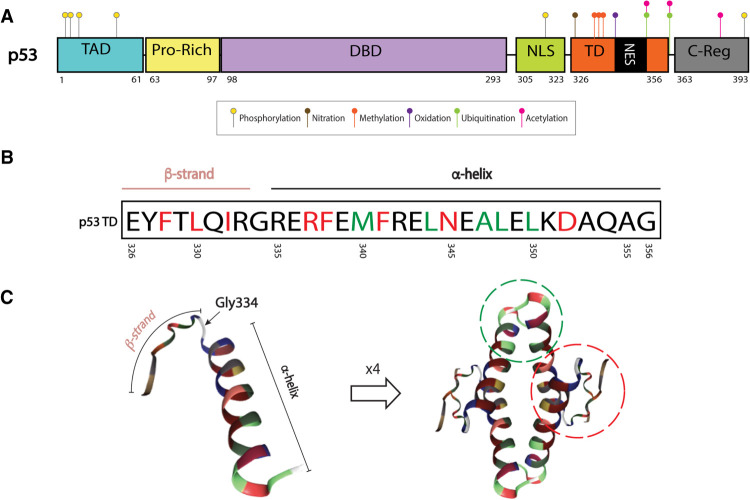
Figure 1.
p53 tetramerization domain (TD) structure. (A) Map of p53 protein. Posttranslational modifications (PTMs) known to be important for tetramerization dynamics and TD-regulated processes: serine 315 and 392 phosphorylation (yellow), tyrosine 327 nitration (brown), arginine 333, 335, and 337 methylation (orange), methionine 340 oxidation (purple), lysine 351 and 357 ubiquitination (green), and lysine 351 and 357 acetylation (pink). PTMs that require tetramerization: serine 6, 9, 15, and 46 phosphorylation (yellow) and lysine 382 acetylation (pink). Ubiquitination in C-terminal lysines (dependent on tetramerization) not shown. (B) The p53 TD is composed of a β strand, a tight turn (Gly334), and an α-helix. Residues involved in dimer and tetramer formation are depicted in red and green, respectively (Chène 2001) (C) Monomer or single peptide (left) and tetramer (right) forms of the p53 TD. Monomer–monomer interactions of β sheets and α helices are depicted by a red dotted line. Dimer–dimer interactions of α helices are depicted by green dotted line (residues outside of these circles can also be involved in stabilizing each complex). Structures extracted from PDB ID 1AIE (Mittl et al. 1998; Berman et al. 2003).