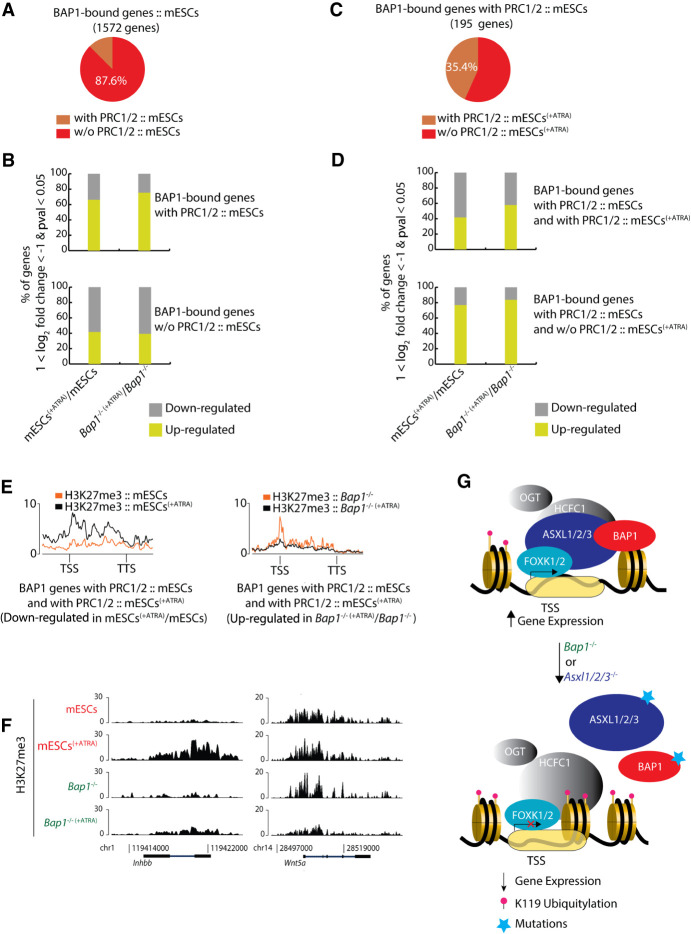
Figure 5.
Correlation between PR-DUB target genes and PRC1/2 reveals the role of PR-DUB in gene expression. (A) Pie chart depicting the percentage of the 1572 BAP1-bound genes, which either colocalize or not with PRC1/2 in proliferating mESCs. (B) The upper panel shows the % of the BAP1-bound genes that are up- or down-regulated in response to ATRA and are also bound by PRC1/2 in proliferating (untreated) mESCs. The lower panel shows the % of the BAP1-bound genes that are not bound by PRC1/2 in proliferating (untreated) mESCs, which are up- or down-regulated in response to ATRA. (C) Pie chart depicting the percentage of the 195 BAP1/PRC1/PRC2-bound genes in proliferating (untreated) mESCs, illustrating the proportion that retain or lose PRC1/2 in response to 72 h ATRA treatment (mESCs[+ATRA]). (D) The upper panel shows the % of the BAP1/PRC1/PRC2-bound genes in mESCs, which retain PRC1/2 in mESCs(+ATRA) and are up- or down-regulated in response to ATRA-induced differentiation. The lower panel shows the % the BAP1/PRC1/PRC2-bound genes in untreated mESCs, which lose the PRC1/2 binding in mESCs(+ATRA) and are up- or down-regulated in response to 72 h of ATRA treatment. (E) The average signal (in metagenes) of H3K27me3 in the wild-type (left) or Bap1−/− (right) mESCs, in untreated (orange) and in response to ATRA-induced differentiation (black) for each cell type, for the BAP1/PRC1/PRC2 bound genes in mESCs which retain PRC1/2 in mESCs(+ATRA) and are either down-regulated (left) or up-regulated (right) upon 72 h of ATRA treatment. (F) ChIP-seq screenshot for two representative loci (based on Fig. 4E) showing the binding of H3K27me3 in untreated and ATRA-treated wild-type mESCs and Bap1−/− mESCs. (G) Model for the role of the PR-DUB complex in regulating histone H2AK119ub1 levels and transcription. BAP1 binding to chromatin depends on FOXK1/2 and the formation of a PR-DUB complex. The binding is required for retaining a chromatin environment that supports gene expression. Loss of BAP1, FOXK1/2, or ASXL1/2/3 leads to loss of PR-DUB from its target genes, the deposition of H2AK119ub1, and decrease in gene expression.