Fig. 5. UHRF1 and DNMT1 maintenance of DNA methylation is important for silencing BL LMP1 and 2A expression in the absence of EBNA2.
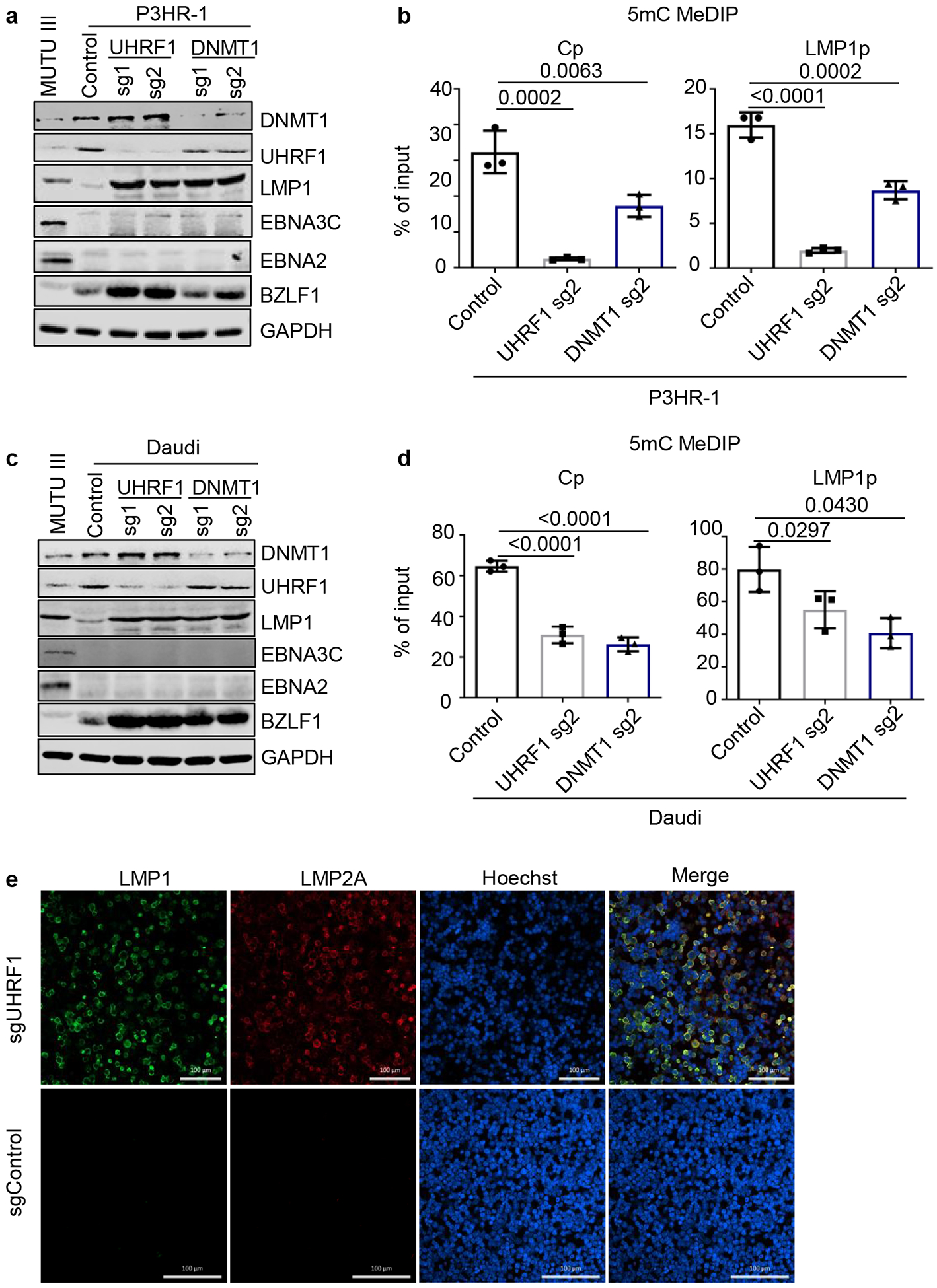
a, Immunoblot analysis of WCL from MUTU III or from P3HR-1 BL that expressed the indicated control, UHRF1 or DNMT1 sgRNAs. P3HR-1 harbor EBNA2-deleted EBV genomes that preclude Cp-activated latency III expression.
b, 5 mC MeDIP was performed on DNA from P3HR-1 BL that expressed control sgRNA (black bars), UHRF1 sg2 (gray bars) or DNMT1 sg2 (blue bars), followed by qPCR for Cp or LMP1p. Mean ± SEM for n=3 biologically independent replicates are shown. p-values were calculated by calculated using one-way ANOVA with Sidak’s multiple comparisons test.
c, Immunoblot analysis of WCL from MUTU III or from Daudi BL that expressed the indicated control, UHRF1 or DNMT1 sgRNAs. Daudi cells harbor EBNA2-deleted EBV genomes that preclude Cp-activated latency III expression.
d, 5 mC MeDIP was performed on DNA from Daudi BL that expressed control sgRNA (black bars), UHRF1 sg2 (gray bars) or DNMT1 sg2 (blue bars) followed by qPCR for the Cp or LMP1p. Mean ± SEM for n=3 biologically independent replicates are shown. p-values were calculated by paired two-sided student’s t-test with equal variance assumption.
e, Confocal immunofluorescence analysis of LMP1 or LMP2A expression in Daudi BL with control or UHRF1 sgRNAs. Nuclei are stained by Hoechst. White scale bar indicates 100 μM.
