Fig. 6. DNA methylation and PRC1-mediated histone ubiquitylation non-redundantly control latent membrane protein expression.
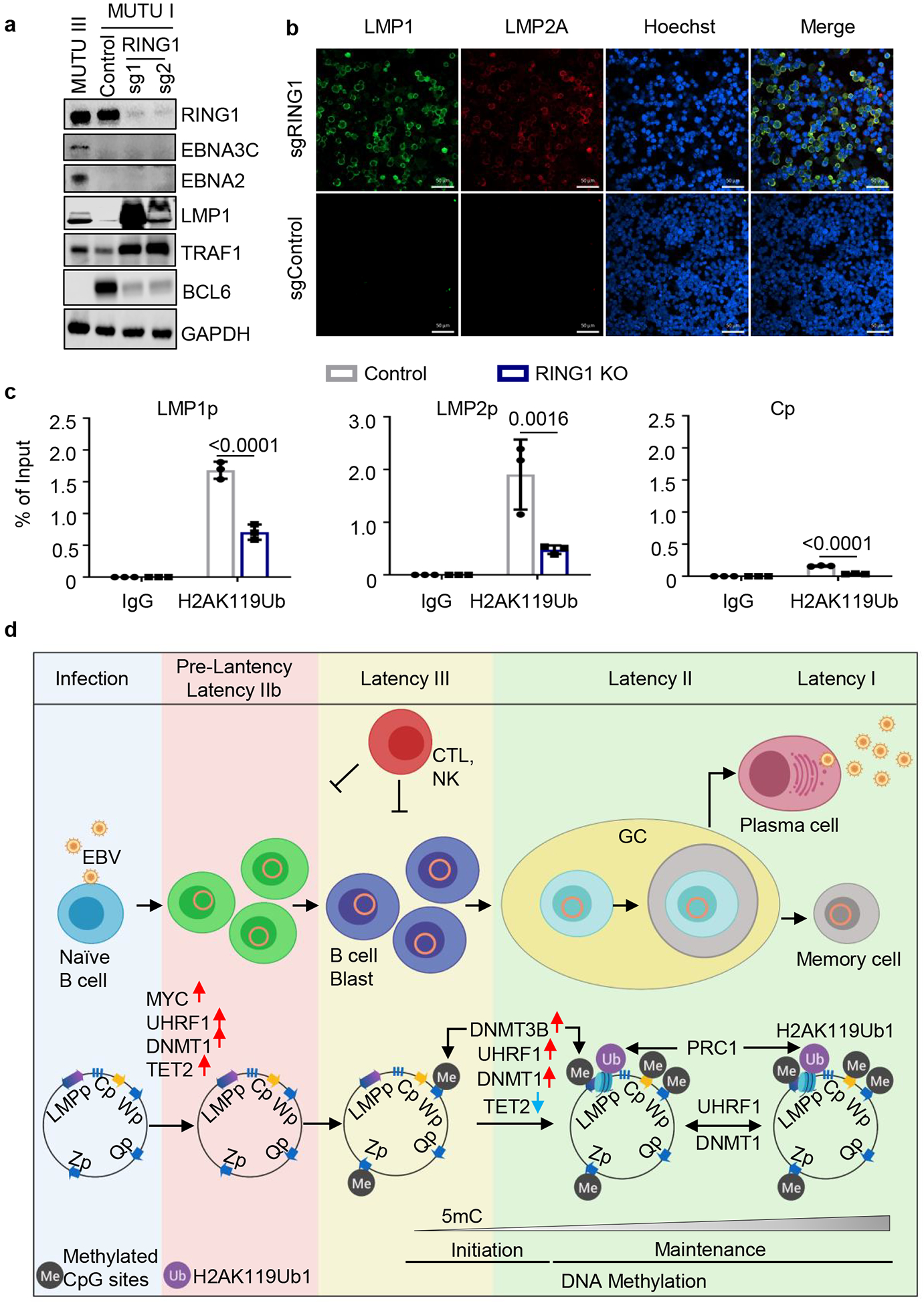
a, Immunoblot analysis of WCL from MUTU III or from MUTU I that expressed control or independent RING1 sgRNAs.
b, Confocal immunofluorescence analysis of LMP1 or LMP2A expression in MUTU I cells with control or UHRF1 sgRNAs. Nuclei are stained by Hoechst. White scale bar indicates 50 μm.
c, ChIP using control IgG versus anti-H2Ak119Ub antibody was performed on DNA from MUTU I with control (black bars) or RING1 (blue bars) sgRNAs followed by qPCR for the LMP1, LMP2 or C promoters. Shown are the mean ± SEM values from n=3 biologically independent replicates. p-values were calculated by calculated using two-way ANOVA with Sidak’s multiple comparisons test.
d, Schematic germinal center model of EBV latency gene epigenetic regulation. Wp and subsequently Cp drive EBNA expression in newly infected cells. UHRF1 and DNMT1 are upregulated early after infection, but EBNA2 also upregulates the TET2 DNA demethylase, which limits DNA methylation. The initiator methyltransferase DNMT3B is not yet expressed, thereby allowing EBV C promoter activation. As MYC levels diminish, LMP promoters are activated and latency III becomes operative. Latency III cells migrate to GCs, where the initiator DNMT3B is induced and where DNMT1 and UHRF1 levels increase. DNMT3b-mediated Cp methylation silences expression of all EBNAs except EBNA1, which instead becomes expressed from Qp. TET2 levels decrease with EBNA2 silencing, further supporting EBV genomic DNA methylation. As infected cells differentiate into memory B-cells, DNA methylation and PRC1-mediated H2AK119Ub1 jointly silence the LMP promoters, allowing transition to latency I, where Qp drives the EBNA1-only program. UHRF1 and DNMT1 maintain DNA methylation in latency I and PRC1 maintains H2AK119UB1, enforcing Cp, Wp and LMPp silencing.
Blots in 6a, b and images in 6e are representative of at least n=3 biologically independent replicates.
