Figure 14:
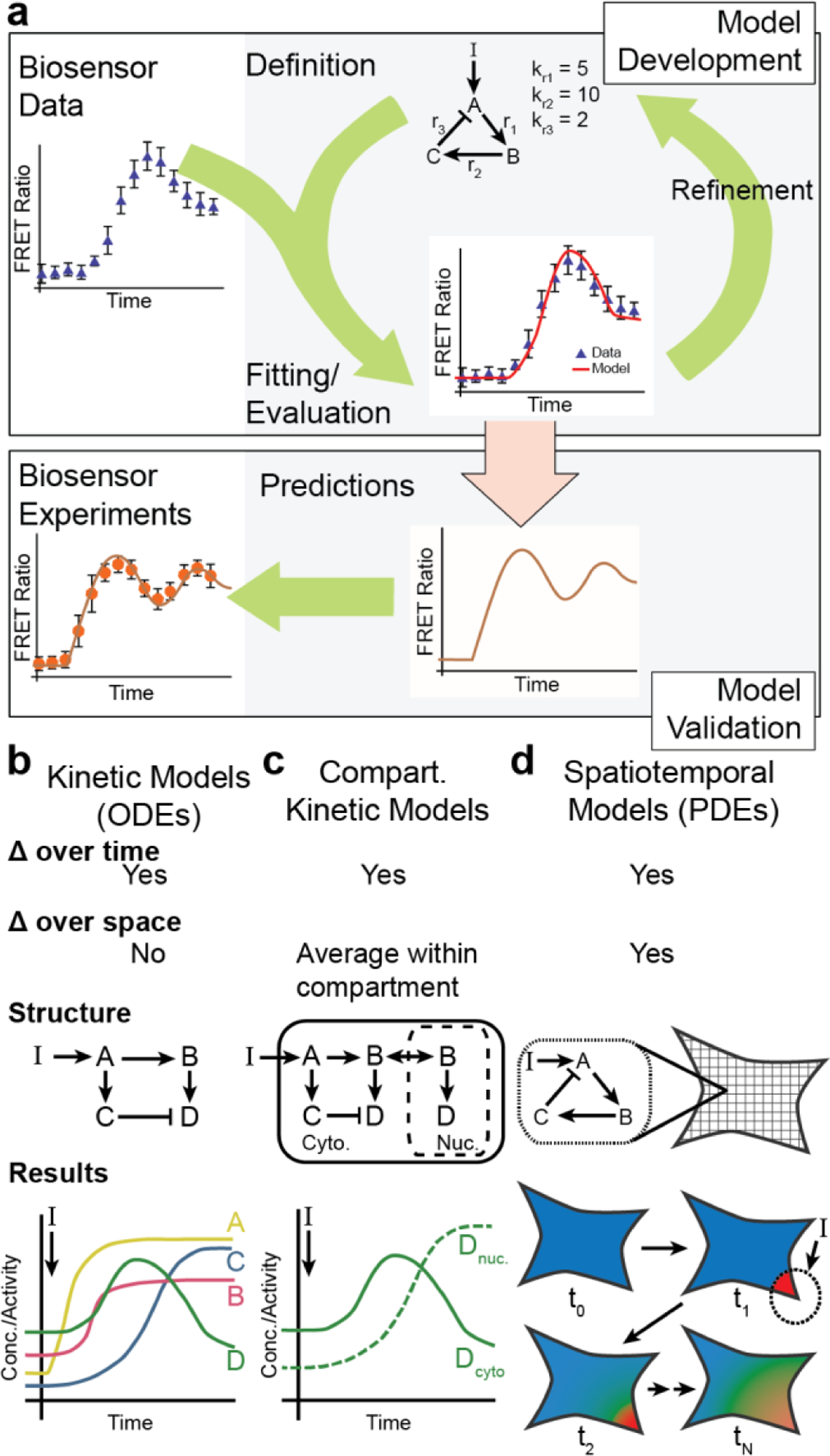
Integrated approach combining biosensor imaging and computational modeling.
A) Computational model development utilizes an iterative approach where the hypothesized structure of the signaling network is implemented into a computational model (top). The results of the computational model are then compared with experimental biosensor data, which serves to approximate unknown model parameters (model fitting) and identify aspects of the experimental data that the model is not capturing. This comparison is then used to refine the hypothesized model structure. This process is iterated until the model adequately reflects the experimental data. This model is then, in turn, capable of generating previously untested conditions (e.g., different stimulation, inhibition of a signaling enzyme) to generate new hypotheses (bottom). Biosensors then serve as a powerful tool to validate these model predictions. B&C) Kinetic computational models simulate changes in the activity and concentration of different signaling reactants as defined by the hypothsized connections within the signaling networks. These models do not directly approximate changes in space (B) but sub-cellular compartments can be defined where specific model species can exchange between compartments (C). D) Spatiotemporal models simulate the changes in signal transduction across both space and time; therefore, the model outputs the model species concentration or activity as it varies across the defined geometry and through time.
