Figure 2:
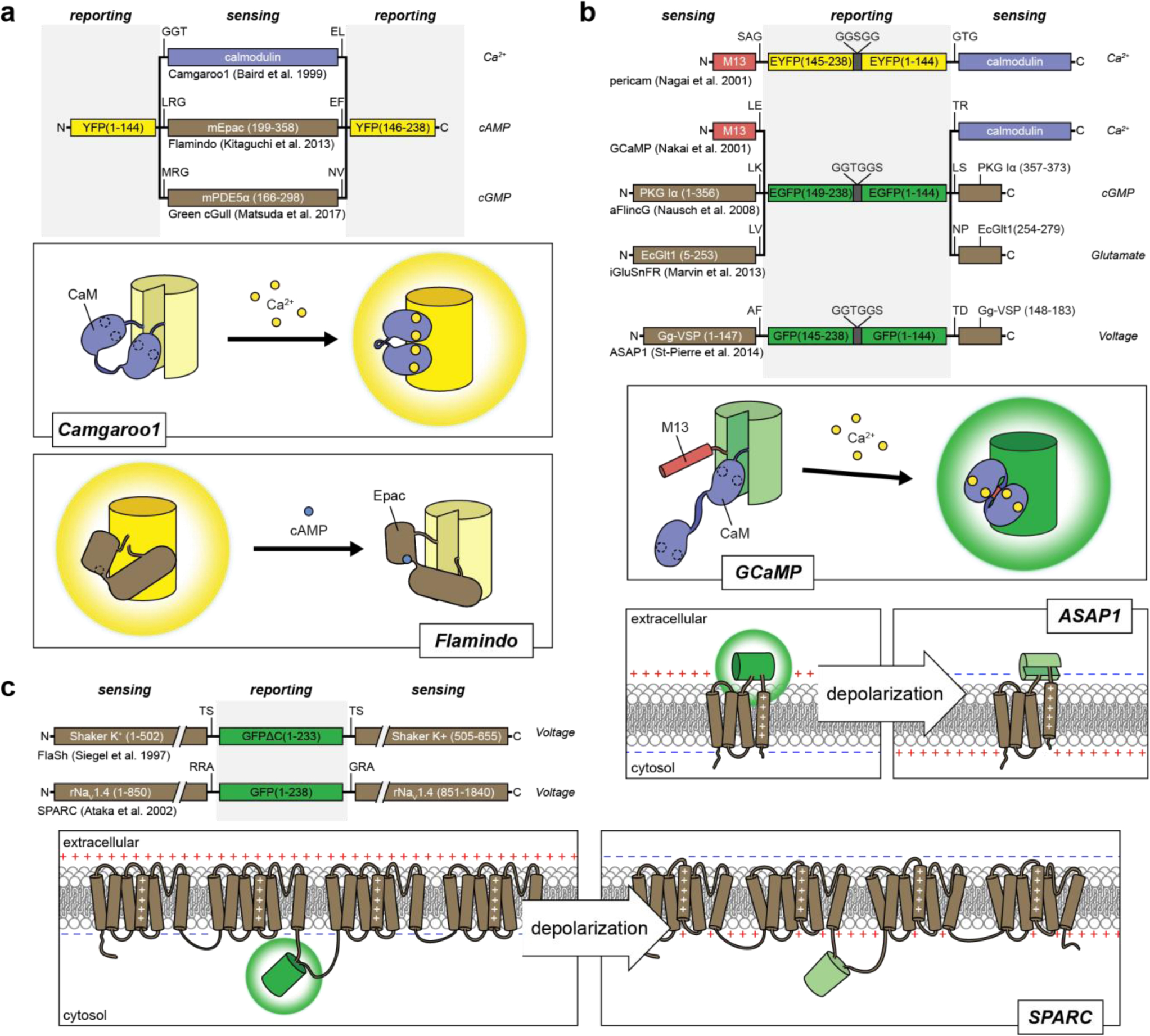
Single FP fluorescent biosensor designs for cellular analytes and membrane potential.
Insertion of a sensing unit into a FP. Calmodulin, mEpac and mPDE5α undergo conformational changes in response to binding Ca2+ 144, cAMP163, and cGMP164, respectively, which perturbs the chromophore and alters the fluorescence. Binding can either lead to an increase in fluorescence, as seen in the Ca2+ biosensor camgaroo1144, or a decrease in fluorescence, as seen in the cAMP biosensor flamindo163. B) Sandwiching a cpFP between sensing units. Biosensors have utilized sensing units that comprise either separate receiver and switch domains148,150 or split proteins that re-constitute during protein folding165–167. For example, GCaMP biosensors utilize separate domains of CaM and M13, where calcium binding to CaM promotes the binding of CaM to M13 and results in a conformational change that leads to an increase in GFP fluorescence. On the other hand, the membrane voltage sensor ASAP1 inserts cpGFP into the voltage-sensing domain of the chicken voltage-sensitive phosphatase Gg-VSP, which reconstitutes after folding and depolarization leads to a conformational change in the 4th transmembrane segment that alters the fluorescence of cpGFP167 C) Insertion of a FP into a voltage-sensitive channel. The conformational changes induced in voltage-sensitive K+ and Na+ channels alter the fluorescence of GFP to act as biosensors of membrane potential168,169.
