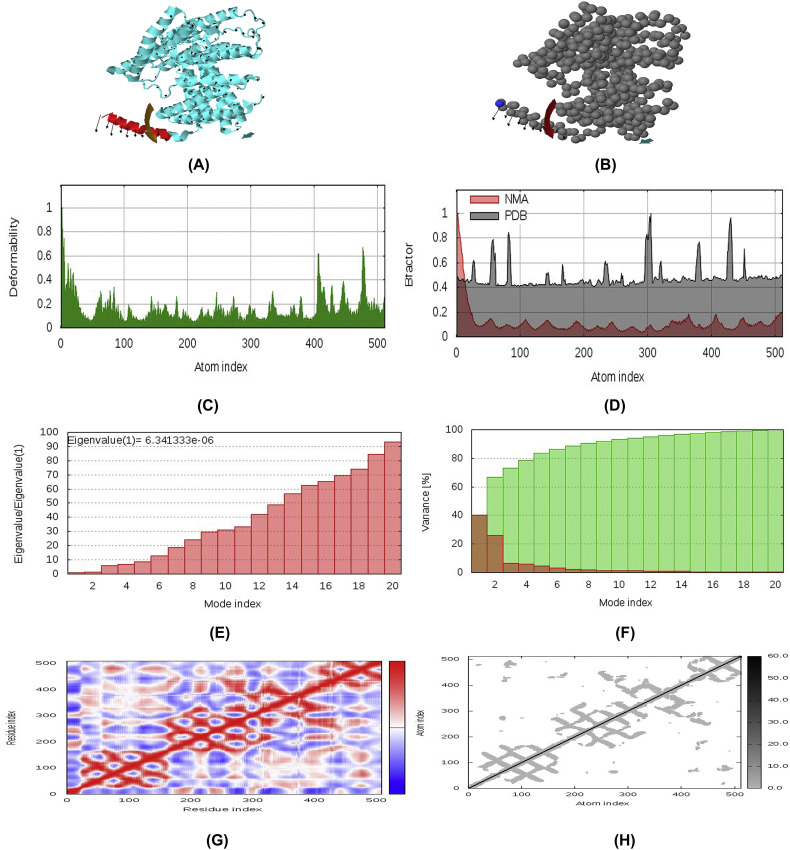
Fig. 10.
Normal Mode Analysis (NMA) of vaccine protein V3. The directions of each residues are given by arrows and the length of the line represented the degree of mobility in the 3D model (A and B). The main-chain deformability derived from high deformability regions indicated by hinges in the chain which are negligible (C). The experimental B-factor was taken from the corresponding PDB field and calculated from NMA (D). The eigenvalue represents the motion stiffness and directly related to the energy required to deform the structure (E). The variance associated to each normal mode is inversely related to the eigenvalue. Coloured bars show the individual (red) and cummulative (green) variances (F). The covariance matrix indicates coupling between pairs of residues, where they may be associated with correlated, uncorrelated or anti-correlated motions, indicated by red, white and blue colors respectively (G). The elastic network model identifies the pairs of atoms connected via springs. Each dot in the diagram is coloured based on extent of stiffness between the corresponding pair of atoms. The darker the greys, the stiffer the springs (H). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)