Figure 3.
OSNs Induce Strong Antiviral Responses upon Infection
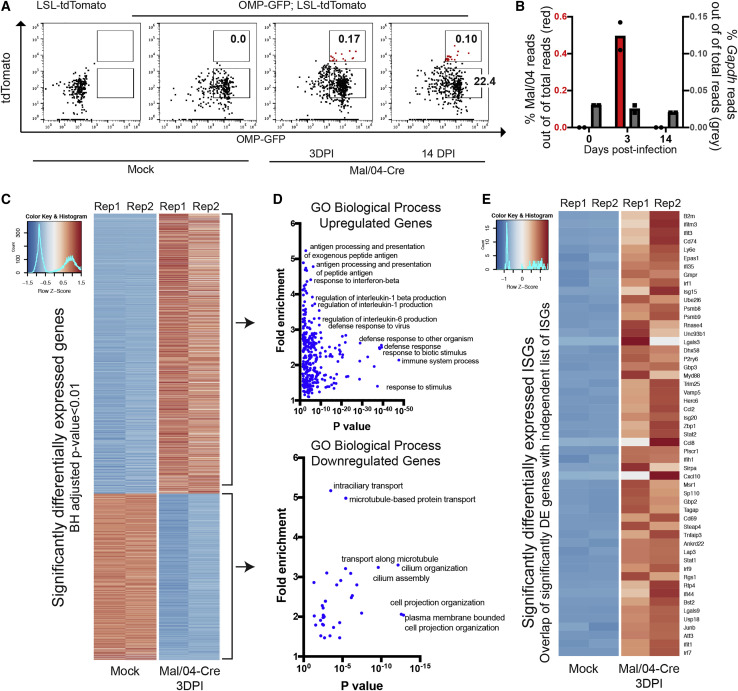
(A) Representative fluorescence-activated cell sorting (FACS) gating strategy to isolate live EpCam+ OMP+ OSNs from the OE of Mock- and Mal/04-Cre-infected mice is shown. For sequencing, tissues from three independent mice per time point were collected and pooled at 0, 3, and 14 days post-infection for mock, actively infected, and previously infected treatment groups, respectively.
(B) Reads mapped to the Mal/04 genome (shown in red) and Gapdh (shown in gray) and normalized to total reads in the Mock (0) and tdTomato+ cell population at 3 and 14 days post-infection.
(C) Heatmap of top significantly differentially expressed genes as ranked by a Benjamini-Hochberg adjusted p value. All “Olfr-” genes were excluded manually prior to analysis to account for sampling variability.
(D) Gene ontology analysis (PANTHER) of both upregulated and downregulated genes in infected OSNs.
(E) Heatmap of differentially expressed genes from an independent list (Schoggins et al., 2014) of interferon-stimulated antiviral genes. Rep, biological replicate.