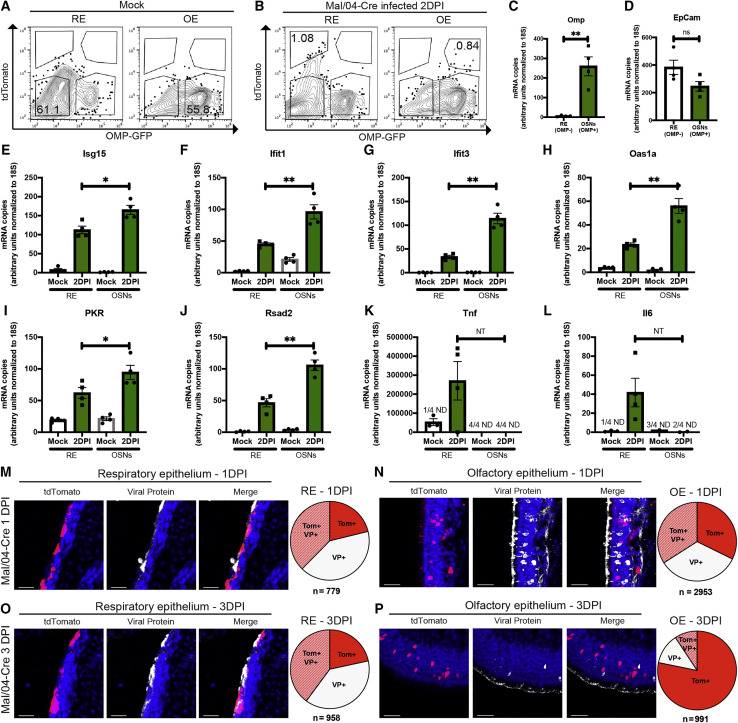
Figure 4.
High-Magnitude Antiviral Responses Are Correlated with Rapid Viral Clearance from Infected OSNs
(A and B) FACS was used to isolate live CD31−CD45−CD326+ cells from the RE and OE of Mock- (A) and Mal/04-Cre-infected (B) mice at 2 days post-infection. Infected OSNs were identified as CD31−CD45−CD326+ OMP+tdTomato+ cells, and infected RE cells were identified as CD31−CD45−CD326+ OMP−tdTomato+ cells.
(C–L) qRT-PCR on transcripts from RE and OSN cells was used to quantify transcripts for (C) Omp, (D) EpCam, (E) Isg15, (F) Ifit1, (G) Ifit3, (H) Oas1a, (I) PKR, (J) Rsad2, (K) Tnf, and (L) Il6. All genes were quantified based on a standard curve and normalized to endogenous 18S expression either in the same sample or samples run on the same plate. Four independent mice were used to collect matching RE and OE samples; a one-way ANOVA with Tukey’s multiple test comparison was used to determine significance. For all graphs, error bars indicate the SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.001. ND, not detected; ns, not significant; NT, not tested due to undetected samples. Data are representative of at least two independent experiments.
(M and N) Microscopy of the RE (M) and OE (N) stained for viral protein from Mal/04-Cre-infected mice at 1 day post-infection. Scale bar: 30 μm. Blue represents DAPI; white represents viral protein; red represents tdTomato. Pie charts represent the number of cells positive for either viral protein only (VP+), viral protein and tdTomato (Tom+ VP+), or tdTomato only (Tom+) collected from 16 images from two independent mice.
(O and P) Microscopy of the RE (O) and OE (P) stained for viral protein from Mal/04-Cre-infected mice at 3 days post-infection. Scale bar: 30 μm. Blue represents DAPI; white represents viral protein; red represents tdTomato. Pie charts represent the number of cells for the indicated groups collected from 24 images from three independent mice.