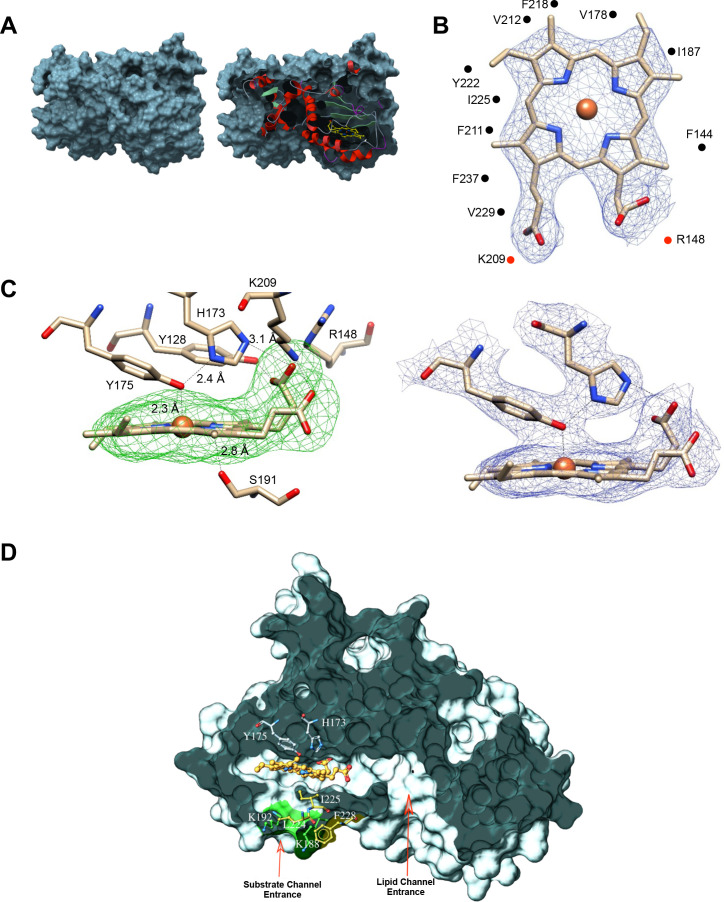
Figure 3. Coordination environment of Sfh5-bound heme.
(A) Left panel shows the surface rendering of Sfh5 in grey. Right panel shows surface view of Sfh5 clipped to expose the buried heme rendered in yellow ball and stick with protein secondary structure elements depicted in cartoon ribbon. (B) Heme bound to chain B of the Sfh5 crystal structure. The 2Fo-Fc electron density map for heme contoured at 1.5 σ is displayed as blue mesh. Residues engaged in van der Waals contacts with heme are shown as gray spheres. The side-chains of residues interacting with the propionyl groups of heme (K209 and R148) are presented as red dots. (C) Left panel: Sfh5 residues that bind heme are shown including the Fe-coordinating residues Y175 and H173. Dashed lines identify the inter-atomic distances between the indicated Sfh5 residues and cognate components of heme. Polder omit electron density map for heme is shown as green mesh (Liebschner et al., 2017), contoured at 2.5 σ. Right panel: Lateral view of the 2Fo-Fc map showing electron density associated with residues coordinating the heme iron center (contoured at 1.5σ). (D) Surface view of Sfh5 clipped to display the ligand-binding cavity. Bound heme (shown in ball and stick and colored by the element with carbons in gold) resides deep inside the cavity. Heme-coordinating residues H173 and Y175 are displayed in ball and stick mode and colored by element with carbons in light gray. Indicated access to the vacant heme coordination site from solvent is potentially controlled by conformational transitions of the K188 and K192 side chains (labeled as substrate channel, entrance displayed in ball and stick render and colored by element with carbons in green), and the side-chains of residues L224, I225 and F228 (colored by element with carbons in yellow). The surface contributed by the side chains of residues K188 and K192 is colored in green. Access channel for lipid to the internal cavity of other Sec14-like PITPs is also indicated.