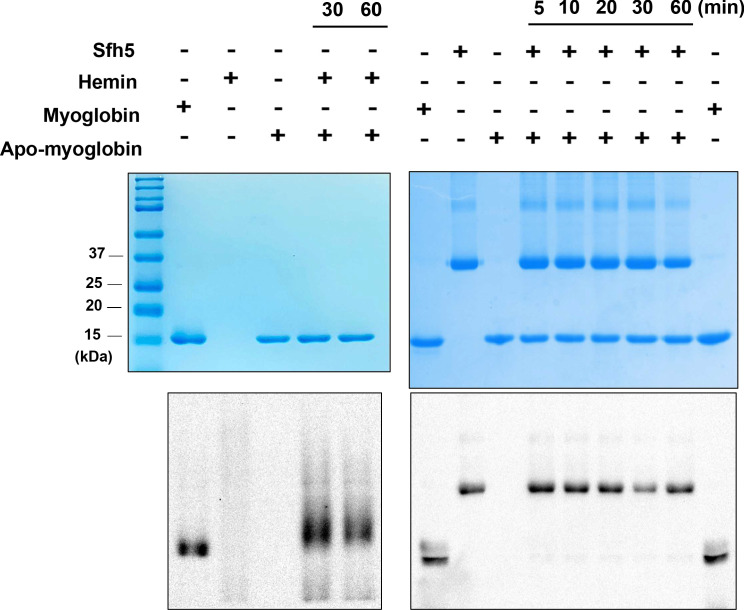
Figure 8. Sfh5 does not donate bound heme to an efficient heme scavenger.
Top panels: Coomassie-stained SDS-PAGE gel images of Sfh5, myoglobin and apo-myoglobin proteins run individually and mixtures of hemin/apo-myoglobin and Sfh5/apo-myoglobin (as indicated) that were pre-incubated for the indicated times. The proteins migrate with the expected masses of 35.7 kDa for the Sfh5 monomer and 18 kDa for myoglobin/apo-myoglobin. Bottom panels show corresponding chemiluminescent images of the nitrocellulose membranes onto which the proteins from duplicate SDS-PAGE gels were transferred and probed by visualizing pseudo-peroxidase activity in situ. Whereas apo-myoglobin avidly scavenged hemin from the medium, no measurable transfer of heme from Sfh5 to apo-myoglobin was detected.