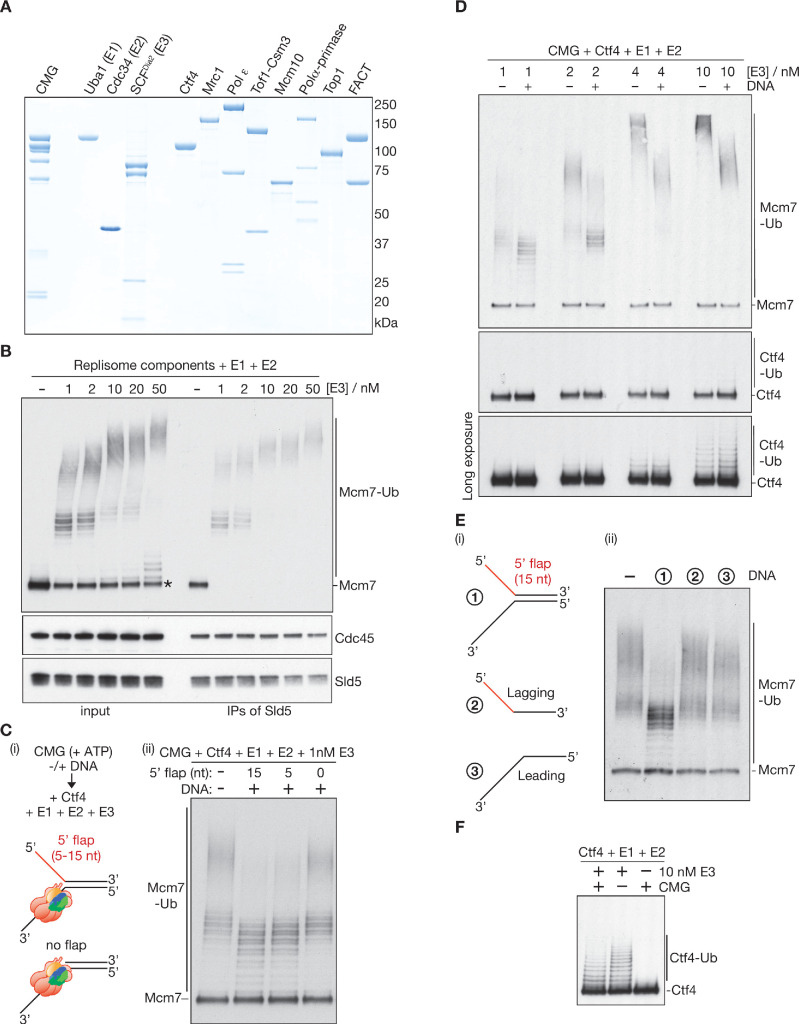
Figure 2. Replication fork structure inhibits CMG ubiquitylation by SCFDia2 and Cdc34.
(A) Purified proteins used in this study, analysed by SDS-PAGE and stained with colloidal Coomassie Blue. (B) The components described in (A) were incubated for 20’ at 30°C with the indicated concentrations of E3 (input), before isolation of CMG (IPs of Sld5) and immunoblotting of the indicated components. The asterisk indicates a small pool of free Mcm7 that was not targeted for ubiquitylation. (C) (i) Reaction scheme and illustration of the association of CMG with model DNA replication forks. Replisome components other than Ctf4 were omitted to limit DNA unwinding by CMG; (ii) Ubiquitylation reactions in the presence or absence of synthetic replication forks with the indicated 5’ flaps. (D) Reactions at the indicated [E3], plus or minus the model fork substrate with 15 nt 5’ flap from (C). (E) Reactions were performed as in (C), with addition of the indicated DNA (2 and 3 are the ssDNA oligos that were used to make 1). (F) Ubiquitylation reactions performed as in (D), in the absence of DNA and +/- E3 or CMG as indicated. See also Figure 2—figure supplements 1–2.
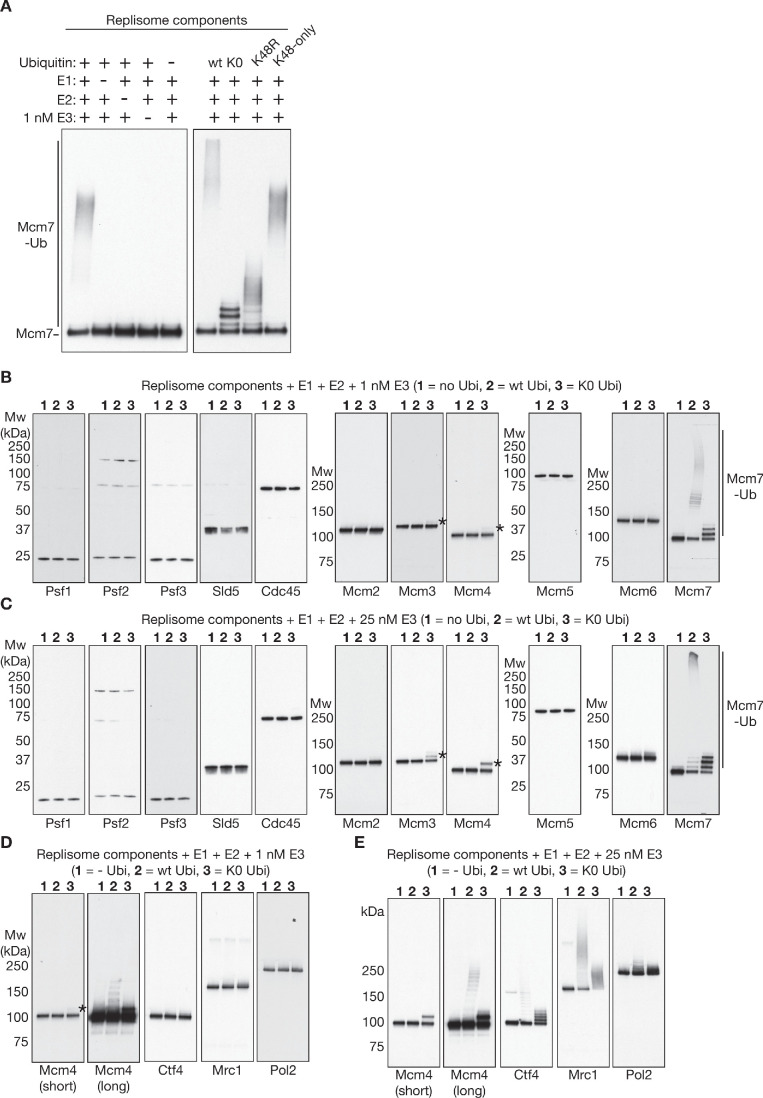
Figure 2—figure supplement 1. Reconstituted CMG ubiquitylation involves the conjugation of K48-linked ubiquitin chains to Mcm7.
Figure 2—figure supplement 2. Association of the CMG helicase with model DNA replication forks.