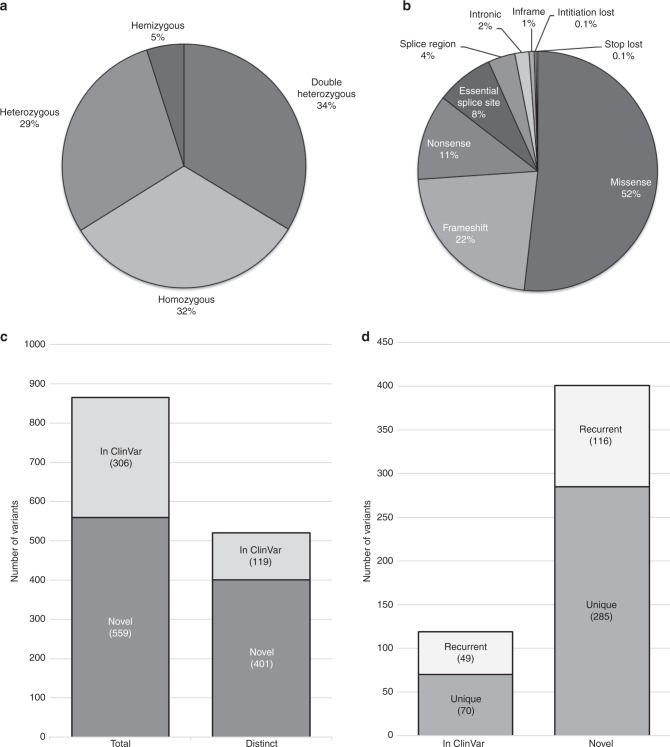
Fig. 3. Breakdown of the suspected pathogenic variants and genotypes in the MYO-SEQ cohort.
(a) Zygosity of the solved patients’ variants (n = 506; 489 patients, 20 with an additional gene to report). (b) Type of variants suspected to be pathogenic. Initiation and stop loss occurred twice each (n = 865). (c) The 865 occurrences were accounted for by 520 distinct variants, of which 119 were reported as pathogenic in ClinVar and 401 were novel in their association to disease at the time of the analysis. (d) Of the 119 distinct variants reported in ClinVar, 70 were detected in only one individual (unique) while 49 were detected in multiple patients. Of the 401 distinct variants that were novel in their association to disease, 285 were detected in only individual cases, while 116 were detected in multiple families.