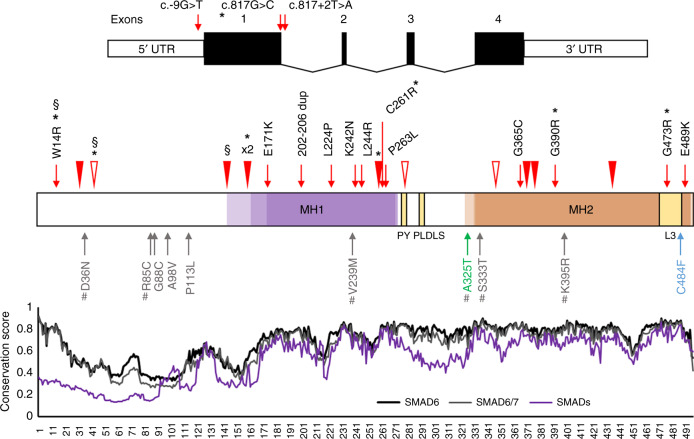
Fig. 1. Human SMAD6 gene and protein showing variants identified in craniosynostosis (CRS).
Top, SMAD6 comprises four exons; positions of variants affecting translation initiation or splicing are indicated. Middle, cartoon of encoded protein showing conserved domains (MH1 and MH2, including highly conserved L3 region) and PY and PLDLS motifs.9 Colored shading indicates the position of the MH1 (transparent purple) and MH2 (transparent orange) domains according to Uniprot, Pfam, and CDD resources, with darker shading denoting overlapping domain assignments. Novel or rare (AFmax < 0.000045) variants identified in CRS patients that are also predicted damaging (loss-of-function [LoF], plus missense variants with DS25 ≥ 4) are indicated above the cartoon, whereas below in gray are additional missense variants predicted to have lower pathogenicity (DS ≤ 3 and/or AFmax > 0.000045); negative and positive controls10 used in the functional assays are colored green and blue, respectively. Frameshifts and stop-gain variants are shown with filled and empty arrowheads, respectively; § = de novo variants; * = novel/rare damaging variants found in addition to the CRS cohort screen; # = AFmax ≥ 0.000045 in gnomAD. Bottom, conservation profiles40 of inhibitory SMADs SMAD6 (black), SMAD7 (gray), and all SMAD members (SMAD1–8) combined (purple line). AF allele frequency, DS deleterious score.