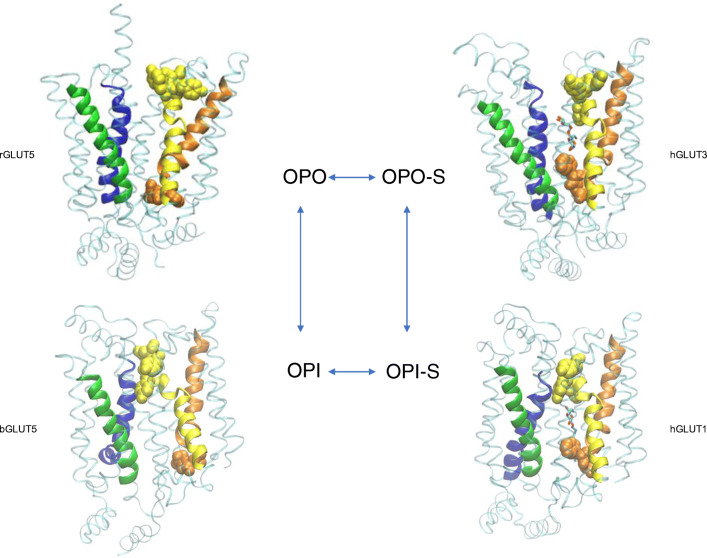
Fig. 2.
Crystal structures of GLUT proteins define the structural basis for alternate exposure of binding site clefts to either outside or inside solutions. Discrete conformations for the GLUTs are identified as OPO (open-outside without substrate; rat GLUT5, pdb:4ybq), OPO-S (open-outside with substrate; human GLUT3 with maltose, pdb:4zwc), OPI-S (open-inside with substrate; human GLUT1 with nonyl-glucoside, 4pyp) and OPI (open-inside without substrate; bovine GLUT5, pdb:4yb9). The structural fold has four inverted trimer repeats with TM1-3 and TM7-9 showing inverted repeat similarity to TM4-6 and TM10-12, respectively. The first TM helix of each of the four trimers is shown as cartoon representation: TM1 (blue); TM4 (green); TM7 (yellow); TM10 (orange), while the rest of the protein is shown as transparent ribbon. The upper half of TM7 (TM7b) and the lower half of TM10 (TM10b) are particularly important for occluding the binding site from the external and internal solutions, respectively. The occlusion OPO to OPI is associated with hydrophobic residues in TM7b moving closer to TM1 while hydrophobic residues in TM10b move away from TM4. The reverse occurs in the OPI to OPO conformational changes. TM7b hydrophobic residues 291, 292, 293 and 294 and TM10b residues 386 and 387 in GLUT1 (with equivalent residues in GLUT3 and GLUT5) are shown with space filling to illustrate this occlusion. In addition, salt bridges between residues at the cytosolic ends of the TMs are formed to bunch these TM ends, and the C-terminal ICH5 region, closer together in the OPO conformations. The location of the substrate glucose moiety that is revealed from both the GLUT3 structures with maltose (OPO-S, and GLUT1 with nonyl-glucose (OPI-S) is the same as in the GLUT3 structure with glucose (not shown, pdb:4zw9). In all cases, the glucose in the central site is polarised with C1-O projecting toward the internal solution with C4-O trailing. The structures shown are constructed using the VMD software and using the pdb files reported and described by the Yan group ([55, 56] for GLUT1, 3) and the Drew group ([158] for GLUT5)