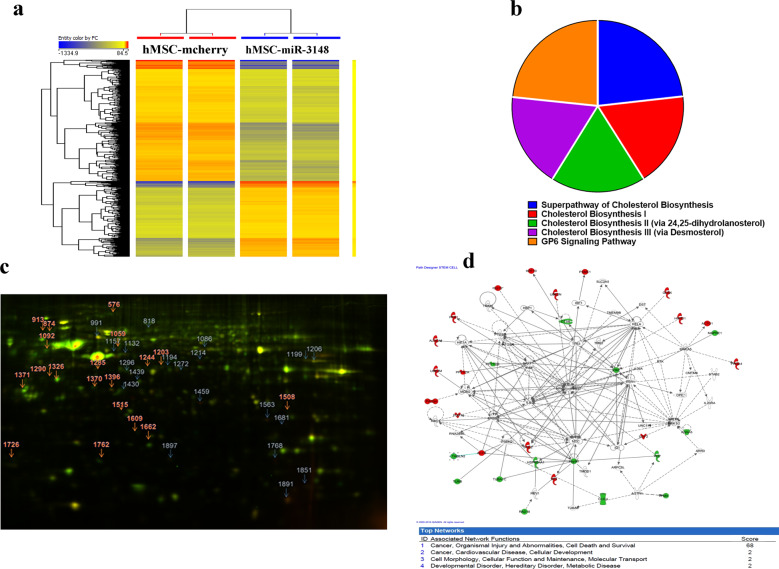
Fig. 2. The hMSC-miR-3148 cell line exhibited multiple altered genetic pathways.
a Hierarchical clustering of hMSC-miR-3148 compared to hMSC-mcherry cells based on differentially expressed mRNA levels as determined by microarray analysis. Each column represents one replica and each row represents an mRNA transcript. The expression level of each gene in a single sample is depicted according to the color scale. b Pie chart illustrating the distribution of the top-5 canonical pathways based on Ingenuity Pathway Analysis (IPA) on the upregulated genes in the hMSC-miR-3148 compared to hMSC-mcherry cells. The size of the chart segment corresponds to activation Z score. c Representative 2D-DIGE gel image. The arrows indicate differentially regulated protein spots between hMSC-miR-3148 and hMSC-mcherry cells as determined by image analysis and identified by MALDI-MS. The gray arrows indicate the upregulated and the red arrows indicate the downregulated proteins in hMSC-3148 cells. d Schematic representation of the “Cancer, Organismal injury and abnormalities, cell death and survival” functional interaction network with the highest score of 68 showing NFkB and IL6 as central nodes. Nodes in green and red correspond to down and upregulated proteins, respectively. Noncolored nodes are proposed by IPA and suggest potential targets functionally coordinated with the differential proteins. Solid lines indicate direct molecular interactions and dashed lines represent indirect relationships.