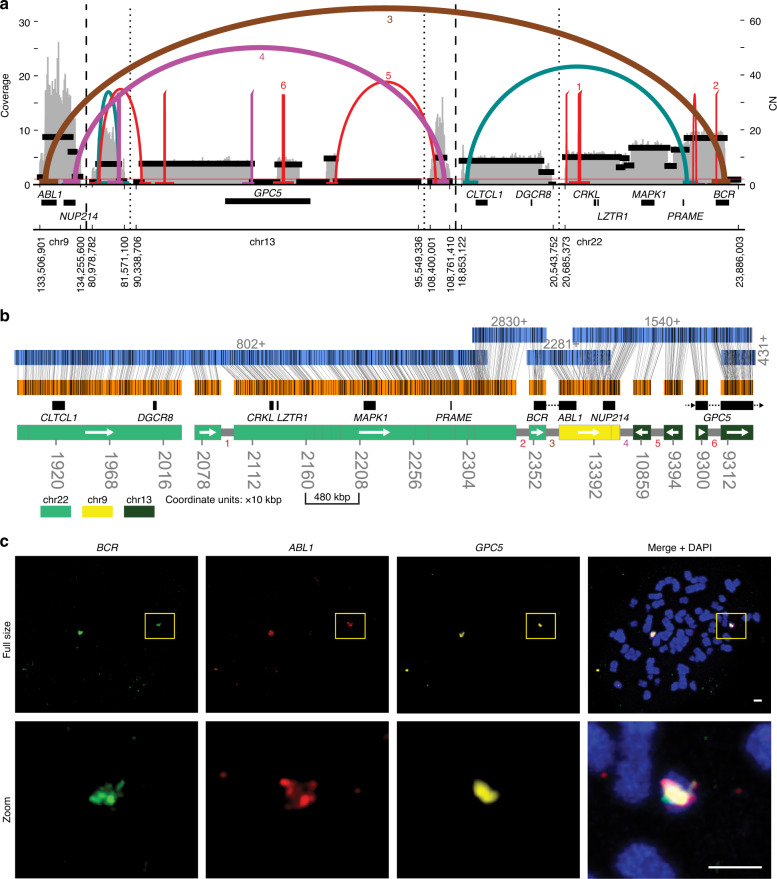
Fig. 3. Reconstruction of a complex Philadelphia chromosome.
a AA-generated breakpoint graph for K562. Estimated copy number (CN), coverage, discordant reads forming breakpoint graph edges, and a subset of the genes in these regions are shown. b AR reconstruction of an 8.5 Mbp focal amplification which was supported by both Irys and Saphyr reconstructions. The tracks from top to bottom are: OM contigs (with contig ID and direction indicated above), graph segments (alignments shown with vertical gray lines), gene subset, and color-coded reference genome bar with genomic coordinates (scaled as 10 kbp units). Gray half-height bars between individual segments on the reference genome bar indicate support from edges in the AA breakpoint graph. White arrows inside the chromosome color bar indicate direction of genomic segment(s). Colored numbers correspond to numbered breakpoint graph edges in panel (a). c Multi-FISH using probes against BCR, ABL1, and GPC5 with DAPI-stained metaphase chromosomes. Scale bars indicate 2 µm in both “Full size” and “Zoom” rows.