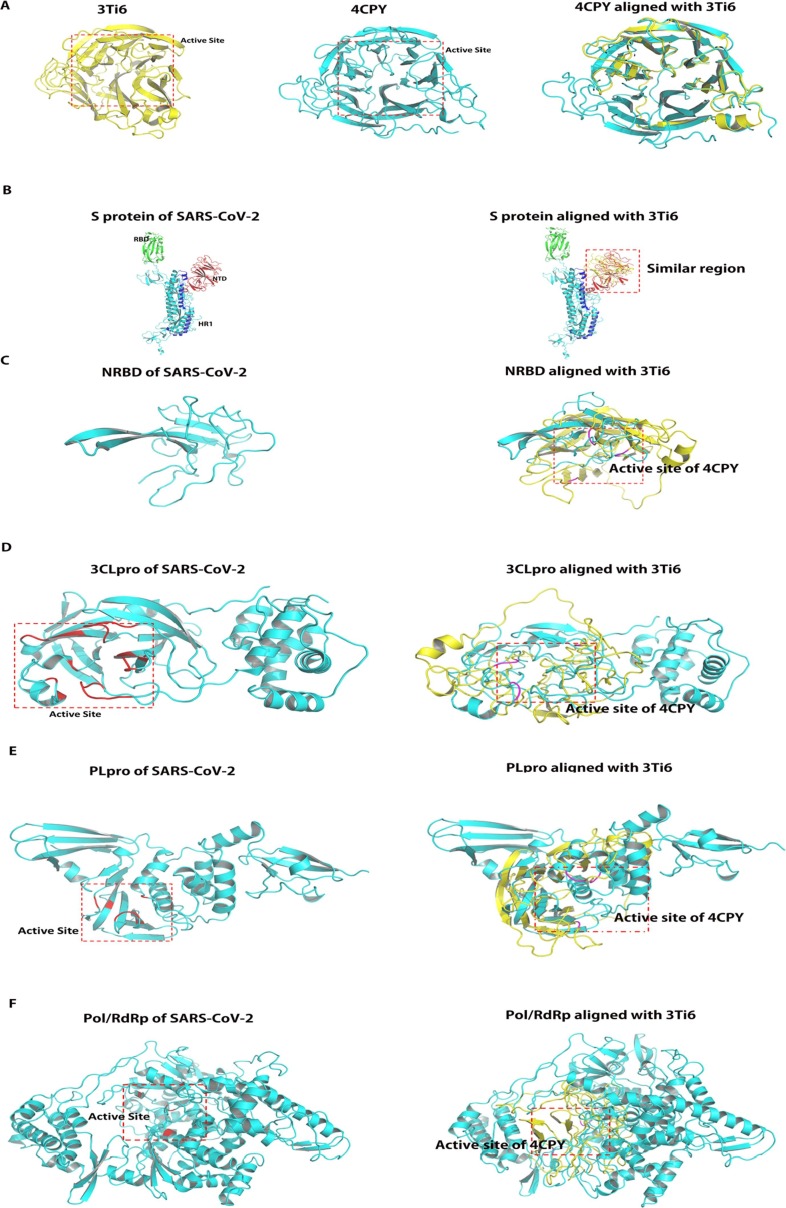
Fig. 2.
Alignment of the structure of viral proteins and influenza A neuraminidase analyzed by TM-align program. (A) Alignment of the structure of influenza A neuraminidase and influenza B neuraminidase. Left panel shows structure of influenza A neuraminidase (PDB ID: 3Ti6) and its active center is framed by red dotted line, Middle panel shows structure of influenza B neuraminidase (PDB ID: 4CPY) and its active center is framed by red dotted line; Right panel: Alignment of 3Ti6 and 4CPY revealed that the active sites of the both structures are the same. (B): Alignment of the structure of S protein and influenza A neuraminidase; Left panel shows structure of S protein and its active sites were RBD domain (Green region) and HR1 (Blue region). Right panel: Alignment of S protein and 4CPY revealed that the active sites of 4CPY is similar with NTD (Red region), suggesting the active sites of the two proteins are not identical. (C) Alignment of the structure of NRBD and influenza A neuraminidase; Left panel shows structure of NRBD, which is the active site of Nucleoprotein. Right panel: Alignment of NRBD and 4CPY revealed that the active sites of the two proteins do not coincide. The key residues of active site of 4CPY are marked in magentas. (D) Alignment of the structure of 3CLpro and influenza A neuraminidase; Left panel shows structure of 3CLpro and its active site is framed by red dotted line, which was based on the key residues marked in red. Right panel: Alignment of 3CLpro and 4CPY revealed that the active sites of the two proteins are similar. The key residues of active site of 4CPY are marked in magentas. (E) Alignment of the structure of PLpro and influenza A neuraminidase; Left panel shows structure of PLpro and its active site is framed by red dotted line. The key residues of active site are marked in red. Right panel: Alignment of PLpro and 4CPY revealed that the active sites of the two proteins do not coincide. The key residues of active site of 4CPY are marked in magentas. F: Alignment of the structure of Pol/RdRp and influenza A neuraminidase; Left panel shows structure of Pol/RdRp and its active site is framed by red dotted line. The key residues of active site are marked in red. Right panel: Alignment of Pol/RdRp and 4CPY revealed that the active sites of the two proteins are not similar. The key residues of active site of 4CPY are marked in magentas. NRBD: N-terminal RNA-binding domain; Pol/RdRp: RNA-directed RNA polymerase; PLpro: papain-like protease; 3CLpro: 3C-like protease; S protein: Spike Protein; RBD: RNA-binding domain; HR1: heptad-repeat 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)