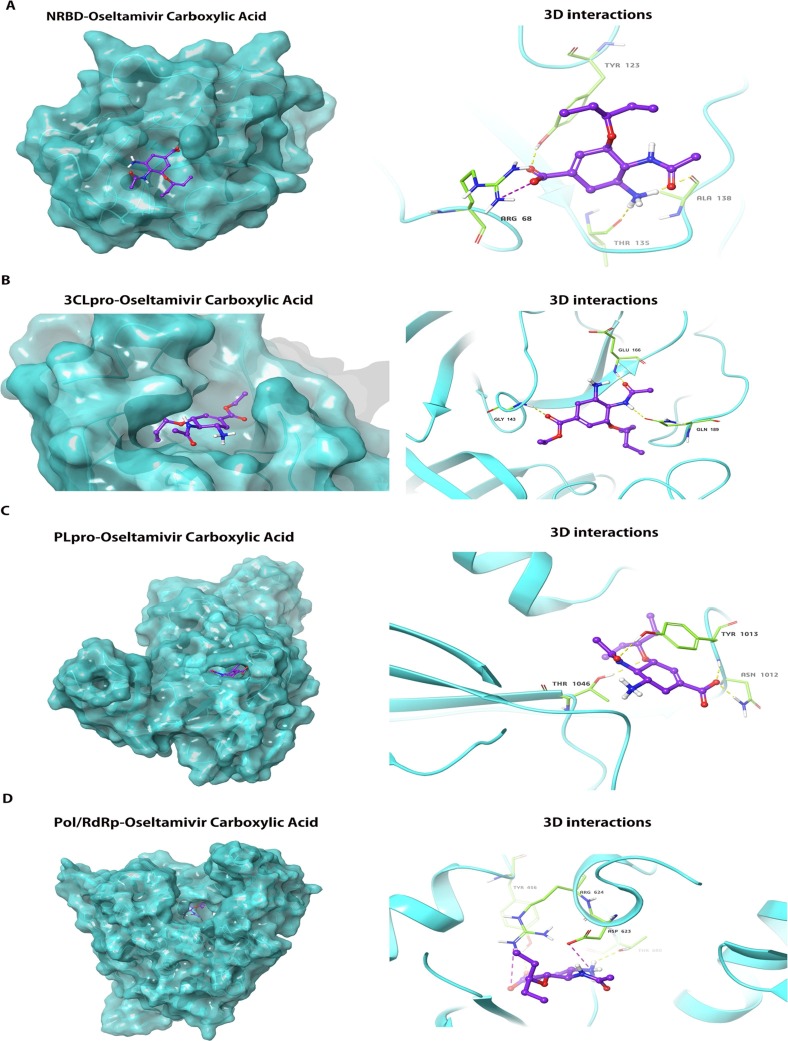
Fig. 3.
The docking conformation and 3D interactions of oseltamivir carboxylic acid inside the active centre of NRBD, 3CLpro, PLpro and Pol/RdRp. (A) Left panel: Docking conformation of NRBD and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of NRBD revealed binding energy of −4.7 Kcal/mol, 3H-bonds(yellow dotted line) and 1 salt bridge(purple dotted line); (B) Left panel: Docking conformation of 3CLpro and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of 3CLpro revealed binding energy of −7.5 Kcal/mol and 3H-bonds(yellow dotted line); (C) Left panel: Docking conformation of PLpro and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of PLpro revealed binding energy of −5.8 Kcal/mol and 4H-bonds (yellow dotted line); (D) Docking conformation of Pol/RdRp and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of Pol/RdRp revealed binding energy of −5.4 Kcal/mol, 2H-bonds (yellow dotted line) and 2 salt bridges(purple dotted line); NRBD: N-terminal RNA-binding domain; Pol/RdRp: RNA-directed RNA polymerase; PLpro: papain-like protease; 3CLpro: 3C-like protease. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)