Figure 4.
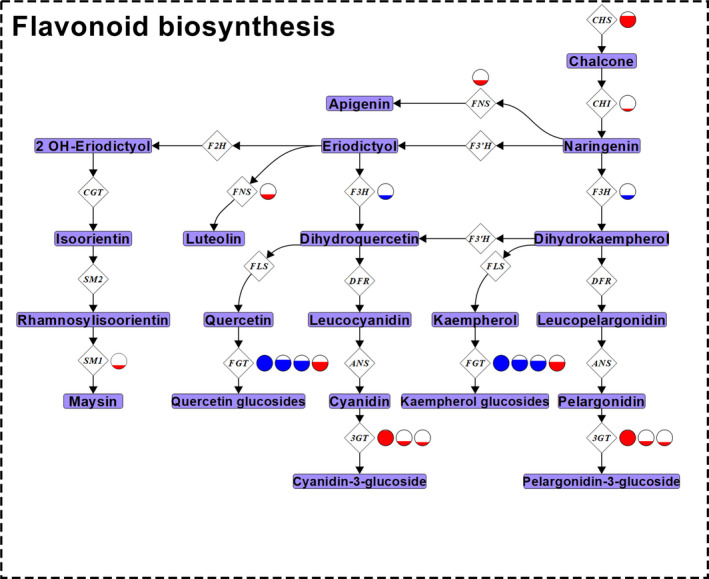
Differential expressed genes between highland and lowland maize landraces encoding enzymes involved in flavonoid biosynthesis. The flavonoid biosynthesis pathway for maize including genes underlying enzymatic steps (white diamonds) and metabolic intermediates (light purple rectangles). Multiple circles next to an enzyme gene name represent multiple genes encoding that enzyme type—cross‐reference Figure 2 and Figure S3 for associated gene names and log2fold change values, respectively. p‐coumaroyl‐CoA, the final product in the general phenylpropanoid biosynthesis pathway (Figure 3), is the entry point into the flavonoid biosynthesis pathway. Anthocyanins—cyanidin and pelargonidin; C‐glycosyl flavones—maysin; O‐glycosyl flavones—apigenin and luteolin; flavonols—kaempferol and quercetin. Genes were considered differentially expressed when significant at a 0.05 FDR rate using a Benjamini–Hochberg correction. Enzyme genes are as follows: CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavonoid‐3‐beta‐hydroxylase; FNS, flavone synthase F3’H, flavonoid‐3’‐hydroxylase; DFR, dihydroflavonol reductase; FLS, flavonol synthase; FGT, flavonol glucosyltransferase; ANS, anthocyanidin reductase; 3GT, flavonoid 3‐O glucosyltransferase; F2H, flavanone‐2‐hydroxylase; CGT, C‐glycosyltransferase; SM2, salmon silk 2; SM1, salmon silk 1. Genes up‐regulated in highland landraces are represented by blue and those up‐regulated in lowland landraces are represented by red. Full circles represent a log2fold change of 2.11 (i.e., 4.32 times higher expression); partially filled circles represent up‐regulation to a portion of 4.32 times
