Figure 4.
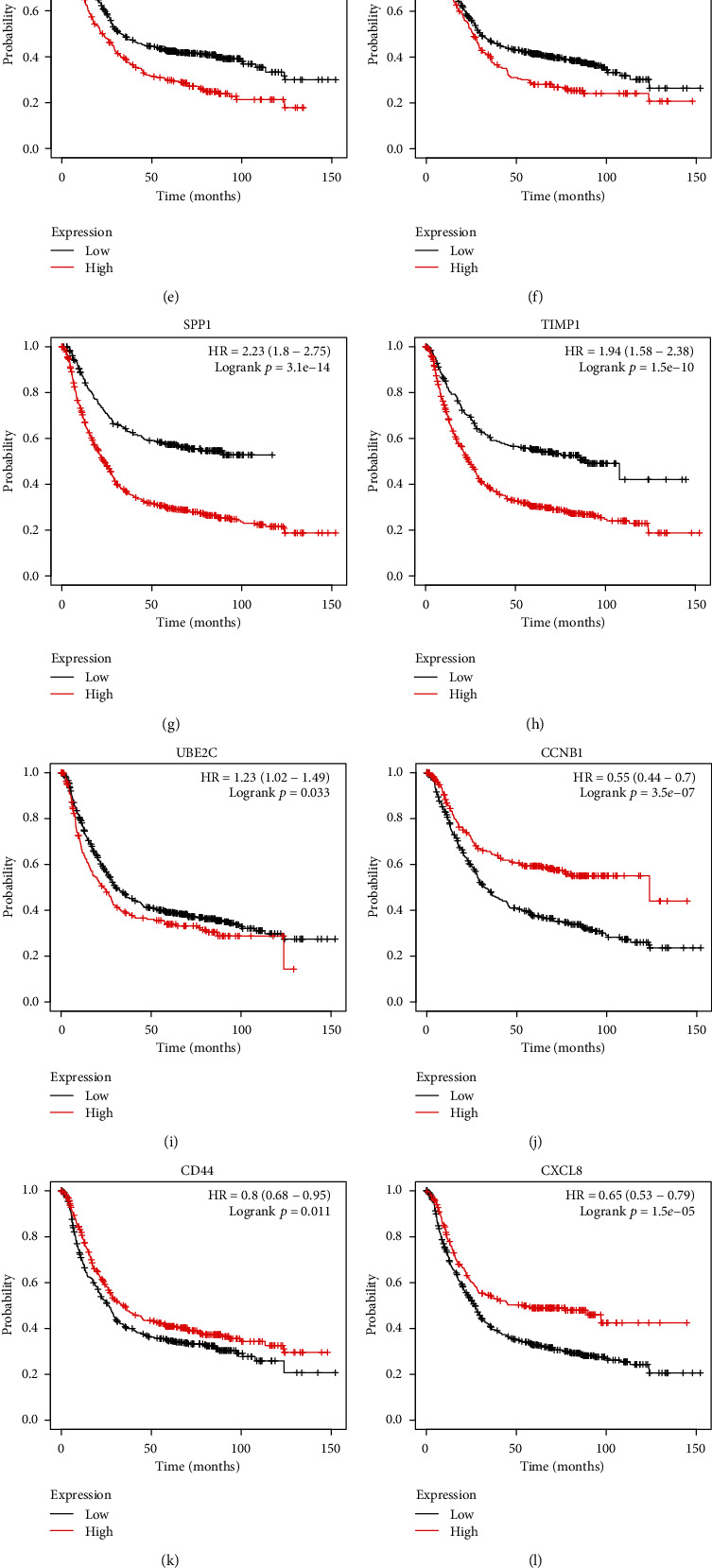
Survival analysis of the top 15 hub genes by the Kaplan-Meier plotter database in GC patient samples. (a–i) Survival analysis of BGN (a), COL1A1 (b), COL1A2 (c), FBN1 (d), FN1 (e), SPARC (f), SPP1 (g), TIMP1 (h), and UBE2C (i) by the Kaplan-Meier plotter database in GC patients. The results show that the survival of GC patients with high expressions of these DEGs was significantly worse (p < 0.01). (j–l) Survival analysis of CCNB1 (j), CD44 (k), and CXCL8 (l) by the Kaplan-Meier plotter database in GC patients. The data show that the survival of GC patients with high expressions of CCNB1, CD44, and CXCL8 were significantly better (p < 0.05). (m–o) Survival analysis of COL3A1 (m), COL5A2 (n), and THBS1 (o) by the Kaplan-Meier plotter database in GC patients. The result shows that COL3A1, COL5A2, and THBS1 were not associated with excessive survival in GC patients (p > 0.05).
