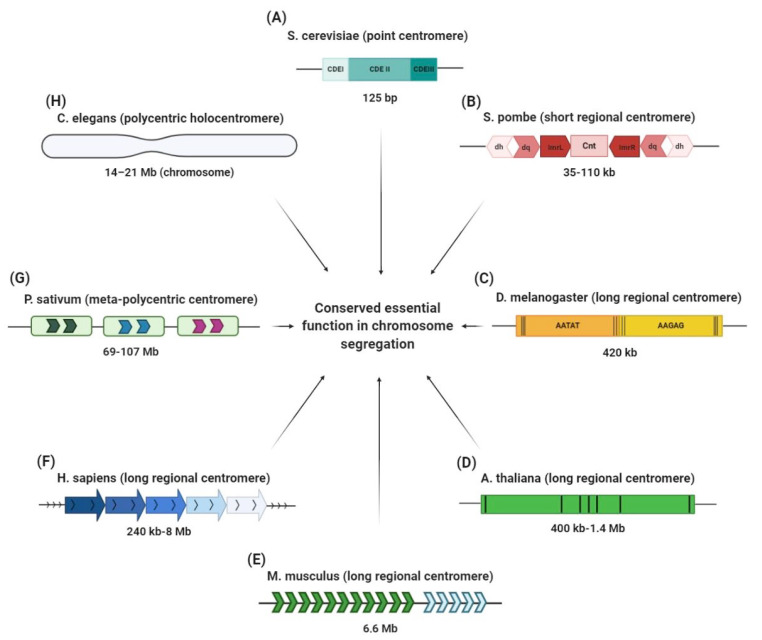
Figure 2.
Centromere structures in different eukaryotes. (A) The S. cerevisiae point centromere is 125 bp in size and it is composed of three centromere DNA elements (CDEs): CDEI, CDEII and CDEIII. (B) The S. pombe centromere is made of inner (ImrL and ImrR) and outer (dg and dh) inverted repetitive sequences that flank a central unique sequence (Cnt). (C) The two main satellite domains (AATAT and AAGAG) of the D. melanogaster centromere are interspersed with transposable elements (black lines). (D) A. thaliana has a 180 bp repeat unit intermingled with retrotransposons (black lines). (E) The mouse centromere is made up of major satellite sequences (MaSat) of 234 bp monomers (spanning ~6 Mb; green arrows) and minor satellite sequences (MiSat) of 120 bp monomers (spanning ~600 kb; blue arrows). (F) Human centromeres contain tandem repeats of α-satellite 171 bp monomers organized head to tail into higher order repeats (HORs). (G) The meta-polycentric centromere of P. sativum is a very long centromere of 13 families of satellite DNA repeats and one family of Ty3/gypsy retrotransposons, organized into 3–5 domains containing CenH3. (H) The polycentric or holocentric centromere of C. elegans covers the entire length of chromosome on which there are several points for microtubule attachment. In spite of this great diversity, all these centromeres perform faithful roles in chromosome segregation.